Research synopsis
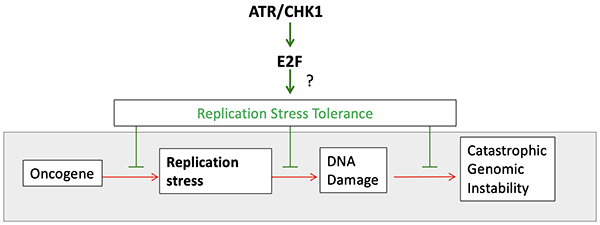
The de Bruin lab's mission is to establish fundamental principles of cell cycle control involved in the maintenance of genome stability and cancer initiation and development. The lab has used a wide range of model organisms and approaches with a final goal of finding ways to identify and exploit therapeutic opportunities for cancer’s addiction to growth and proliferation. Our group has established the molecular mechanism by which G1/S transcription is inactivated during S phase and how it is maintained in response to replication stress. Our work indicates that G1/S transcription plays an essential role in the tolerance to replication stress. Since replication stress is a major cause of genomic instability in cancer, this represents a crucial new area for cancer studies in the lab.
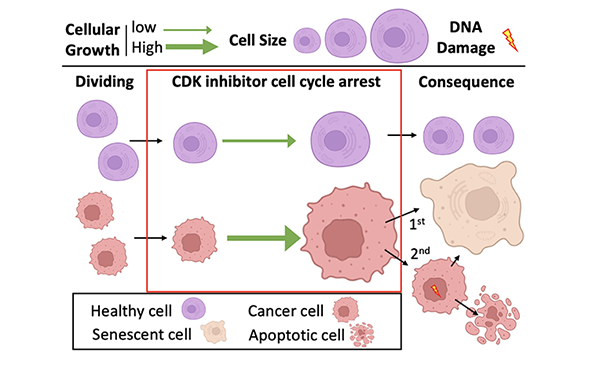
 We recently established that cellular growth has a central role in determining CDK inhibitor sensitivity. This work suggests that independent of increased CDK activity, when cell division is inhibited, but cell growth continues, cells simply get too big to maintain a proliferative state or maintain genome stability. Since enhanced growth signalling is common in cancers, this helps explain the anti-cancer efficacy of CDK inhibitors. This new insight is now being used to ask more specific questions to understand the mechanisms that determine a cell’s sensitivity to CDK inhibitors.
We recently established that cellular growth has a central role in determining CDK inhibitor sensitivity. This work suggests that independent of increased CDK activity, when cell division is inhibited, but cell growth continues, cells simply get too big to maintain a proliferative state or maintain genome stability. Since enhanced growth signalling is common in cancers, this helps explain the anti-cancer efficacy of CDK inhibitors. This new insight is now being used to ask more specific questions to understand the mechanisms that determine a cell’s sensitivity to CDK inhibitors.
Selected publications
Peripolli S, et al (2024). Oncogenic c-Myc induces replication stress by increasing chromatin occupancy of cohesins at CTCF sites. Nature Communications, in press.
Wilson GA, et al (2023). Active growth signalling promotes senescence and cancer cell sensitivity to CDK7 inhibition. Molecular Cell.
Manohar S, et al (2023). Genome homeostasis defects drive enlarged cells into senescence. Molecular Cell.
Matthews H, et al (2021). Cell cycle control in cancer. Nature Reviews Molecular Cell Biology.
Pennycook B, et al (2020). E2F-dependent transcription determines replication capacity and S phase length. Nature Communications.
Bertoli C, et al (2013). Control of cell cycle transcription during G1 and S phases. Nature Reviews Molecular Cell Biology.
 Close
Close


