+44 (0)20 7679 7271
LMCB Room 3.11
Research synopsis
The primary interest of our laboratory is to understand how cells make decisions about their identity during development. Despite the intense activity in the field of developmental biology for many decades, our understanding of how cells make decisions is decidedly limited. Models tend to fall into two categories— instructive or permissive. Instructive models emphasise the external or inherited trigger, giving the cell no choice. Permissive models view the cell as waiting for some event that triggers a decision already made, often incorporating a central “noise plus feedback” element. These models can be useful in the limited number of apparently simple contexts in which cell fates appear to be determined by a limited number of components, yet in most systems, which are neither wholly instructive nor permissive, and often depend on a highly complex set of interactions between the cell and its environment, these opposing models lack utility.
To allow a deeper understanding of cell decision making, and the generation of more useful conceptual frameworks, we have developed and implemented a range of technologies- with the view that to understand the decision of cell requires monitoring a broad range of regulatory features of single cells, as they differentiate. These approaches encompass both imaging and transcriptomic methods for monitoring the signalling (see Movie 1 below) and gene expression dynamics of single cells. In particular, we use imaging to directly observe the bursts of activity of individual genes in living cells (see Movie 2 below). This approach provides a real time readout of the gene expression decisions of the cell, in parallel with other features of the cells and their environment. We combine these techniques with detailed quantitative analysis of the data, and use modelling and molecular genetics to refine our hypotheses.

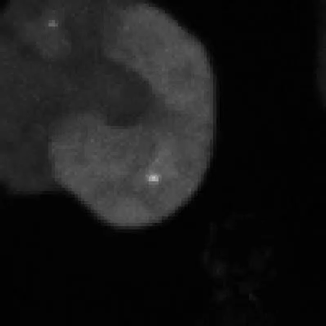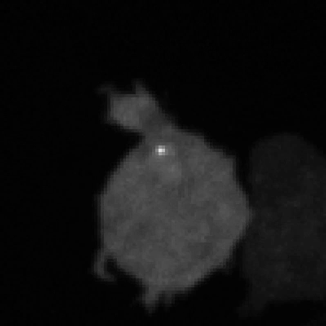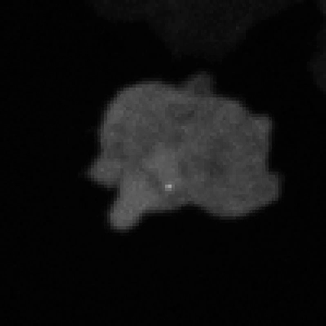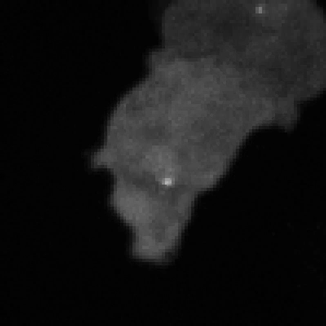
Legend for movies | Left: dynamics of long-range signalling in cell populations. Movie shows periodic cAMP signalling waves spreading through populations of Dictyostelium cells. Right: Fluctuations in the transcriptional activity of a single gene in living cells. Newly synthesised RNA, from a gene encoding actin, is visualised as a fluorescent spot using the MS2 labelling technique. Movies such as this reveal the “bursts” or “pulses” of gene firing.
 Close
Close



