Research synopsis
In every cell, growth, proliferation and survival are key processes that require tight regulation by signal transduction pathways. Defects in this regulatory control can lead to severe diseases, such as cancer.
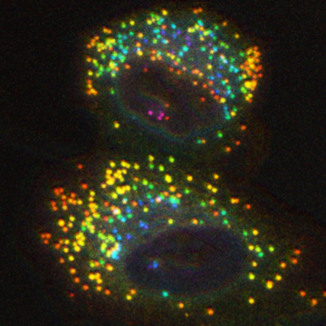
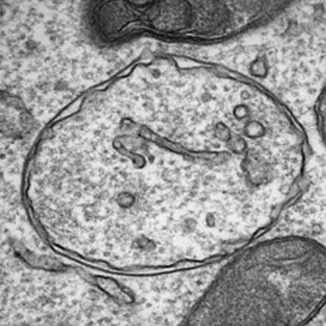
Cells maintain a balance of growth and survival by two main processes: Growth factor signalling (under nutrient-rich conditions) and autophagy (under nutrient-scarce conditions). Over the past years, it has become clear that these processes are tightly co-regulated.
In my lab we aim to elucidate the molecular interactions between these pathways. We use systematic genomic approaches to identify networks of cell signalling and autophagy. We employ high-throughput screening technologies as a means to identify novel components that modify cellular signal transduction pathways and autophagy. To this end, we are developing novel screening technologies based on arrayed CRISPR libraries and using knockout cell panels.
Our studies will generate strategies for the identification of potential therapeutic targets in diseases such as cancer and neuro-degeneration. Further, we have the resources and expertise to translate these basic research findings into a drug discovery program to identify potential therapeutic lead compounds.
Selected publications
About the lab
Funders
Research themes
Technology
People
Collaborators
 Close
Close


