(1) My spectrum was not received by e-mail - It is likely that space was left in the dataset name. For example, the log file shows the following error if the data set is named "abc 20 jan":
"Couldn't send email to somebody@something.com
Error: 400: couldn't open "{C:/Bruker/TOPSPIN/data/xyz/nmr/abc 20 jan/10/pdata/1/abc 20 jan_10_1.pdf}": no such file or directory"
Replacing "abc 20 jan" with ""abc_20_jan" would be sufficient. As a general rule, however, avoid using long names with special characters and spaces. Dataset names with 8 characters consisting of letters, followed by "_" or "-" and then numbers are acceptable (e.g., ABC1001 or Abc-1001 or abc_1001).
(2) My sample fails to run - If "." is left at the end of the dataset name, then the program is likely to fail. Avoid using spaces and special characters, such as ". , : ; $ £ | \ / ? ! > < ¬ ^ * ` % + = ~ # & @ ( )". See above for correct examples of dataset names.
(3) My spectrum appears upside down or poorly phased. Do I need to rerun it? - There is no need for rerunning such a spectrum and it can be phased correctly using NMR software. If using TopSpin, type ".ph" to enter phasing mode, press "0" button on the top left of the window and keep it pressed while moving the mouse. This will allow to correct the phase of the peak centred on the vertical red line. In a similar manner correct phases of other peaks using the "1" button". Type ".sret" or click on the floppy disk button to save the phase of the spectrum. If phases appear to be correct, but the spectrum is upside down, then typing "nm" under TopSpin would revert it.
(4) I entered a wrong solvent when submitting my sample - As above, there is usually no need for a rerun. Chemical shifts can be recalibrated using NMR software. If, for example, CDCl3 was selected wrongly instead of DMSO, look for a characteristic DMSO signal at 7.26 ppm in your proton spectrum and change its chemical shift to 2.50 ppm. Under TopSpin, type ".cal" to enter the calibration mode in order to change the chemical shift of the residual solvent peak. Take notice of the SR value and use it in 2D spectra, if any.
(5) My 2D spectrum failed to run - This usually happens when 1D projections in F2 and F1 dimensions are not available or they are wrongly defined on sample submission. In the example below, the 2D spectrum for the sample in position 31 will run without errors while that for the sample in position 32 will fail:
(6) I have installed TopSpin under Linux, but compiling au programs fails - Various solutions may exist, but the following two solutions may also work for your Linux installation.
Under Centos 7, installed TopSpin 3.2 without gcc, uncommented line 20 in makeau and installed glibc-devel:
cd /opt/topspin/exp/stan/nmr/au
vi makeau ;uncomment line 20
yum install glibc-devel
Under Linux Mint 17, installed TopSpin 3.2 with gcc 4.5.2. On compiling an au program, "error: sys/cdefs.h: No such file or directory" was issued. After additional installation using:
apt-get install libc6-dev-i386
another error occurred: "asm/errno.h: No such file or directory". Installed linux headers:
sudo apt-get install linux-headers-generic
This installed headers for 3.13, whereas the installed version was 3.18, but this does not matter in this case. Then:
cd /usr/include/
cp -pR asm-generic/ asm/
Compilations of au programs proceed without errors after the above.
(7) I would like to run N spectra and e-mail each spectrum to a different user - Iconnmr allows to import a spreadsheet file. Create a file with a csv extension using Excel or a text editor:
Holder Position,Experiment,Solvent,Data Set Name,Mail Data/Notify to
1,PROTON.ucl,CDCl3,AB01,zzzzzz@ucl.ac.uk
2,PROTON.ucl,CDCl3,CD23,yyyyyy@ucl.ac.uk
....
Under Iconnmr, click on File->Import Spreadsheet .csv file. Select the file you created and specify columns needed:
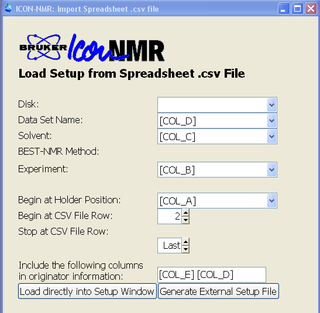
Click on "Load directly into Setup Window".
(8) I would like to run my sample in a solvent without deuterium (e.g. H2O instead of D2O) - It is possible to run spectra in protonated solvents in automation. Please ensure that the solvent is set to "None", which here refers to any solvent that does not contain deuterium. The selection of "None" instructs the shimming routine to use the solvent 1H signal for field optimisations. Then, in the "Par" column click on a dark blue square with "=" in the centre and set LOCNUC to "off" (usually it is set to "2H"). You could run all experiments in this manner, by changing solvent to "None" and LOCNUC to "off". However, a normal proton NMR spectrum will be dominated by the solvent peak(s). Use WATER.ucl under Iconnmr to run samples in solvents with one peak in the NMR spectrum (e.g. H2O or CHCl3). The intensive solvent peak will be suppressed in this experiment. Similarly, you could use DoubleSolventSuppression.ucl and TripleSolventSuppression.ucl, which will suppress two and three biggest peaks, respectively, in the 1H NMR spectrum.
(9) I tried to export an image file using plot0 of TopSpin version 3.2 but it failed with an error about gswin32c.exe and GHOSTSCRIPT_DIR? - To get "export" of plot0 to work under TS3.2/Windows 10, download and install Ghostscript 9.16 for Windows (32 bit) from http://www.ghostscript.com/download/gsdnld.html. Copy 4 files, including gswin32c.exe, from C:\Program Files (x86)\gs\gs9.16\bin into C:\Bruker\Topspin\Gnu\bin. Then, to set environment variable GHOSTSCRIPT_DIR, go to Control Panel → System and Security → System → Advanced system settings → Environment variables and set it to C:\Bruker\Topspin\Gnu\. Restart TopSpin.
(10) I have a sample with very high salt concentration and the 600 MHz fails at the automatic tuning/matching (ATM) stage. How do I disable ATM? - See the screenshot below. You will need to click on the "lock" symbol with blue and red halves first.
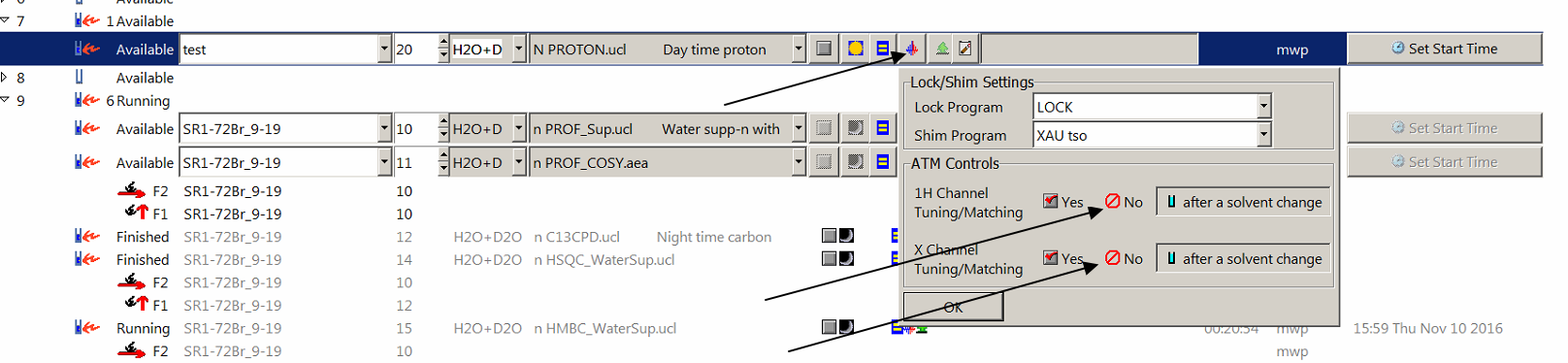
Then click on "No" against 1H and X channels. For important samples requiring a series of experiments, tuning/matching can be adjusted manually by the NMR staff.
 Close
Close

