This initiative focused on the annotation of cardiovascular disease-relevant proteins and microRNAs and was funded by several British Heart Foundation grants from November 2007 to July 2018.
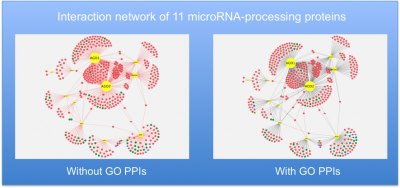
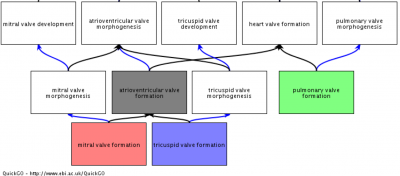
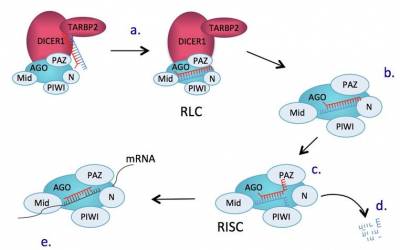
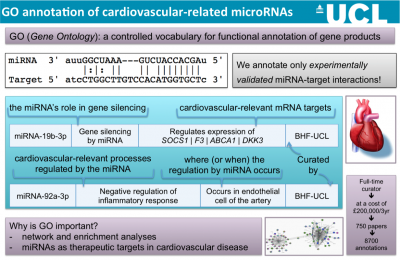

To provide the GO annotations needed to support the cardiovascular research community we have undertaken several focused annotation projects. These processes were prioritised, following discussions with experts in the field and are considered to have important roles in a variety of cardiovascular diseases. GO annotations have facilitated the detailed and high-quality annotation of over 3,600 cardiovascular-relevant human gene products, providing over 32,000 GO annotations, and there are many ways in with the impact of our curation on data analysis and annotation resources can be demonstrated. These annotations were funded by BHF grants (SP/07/007/23671, 2007-2012; RG/13/5/30112, 2013-2018). Annotations contributed by this project to the GO Consortium resource and the IMEx dataset are attributed to BHF-UCL.
The full list of BHF-funded gene products prioritised for annotation, the annotations we have submitted and the Gene Ontology terms we have created are available:
- Cardiovascular-disease-relevant gene products prioritised for annotation
- GO annotations resulting from BHF-funded projects
- GO terms resulting from BHF-funded projects
- Heart development GO terms resulting from BHF-funding
For detailed information about creating GO annotations and GO terms and the overall curation process follow the link below:
To find out more about the heart and cardiovascular system, please see the British Heart Foundation website.
What we curate
- Proteins - GO annotations for proteins to the GO Consortium
- microRNAs - GO annotations for microRNAs to the GO Consortium
- Protein-Protein Interactions - Protein-protein interaction data to the IMEx Consortium
- Foam cell differentiation
The goal of this BHF-funded project was to capture biological information about foam cell differentiation. The list of priority proteins (listed below) were selected from a review of the literature. The annotations associated with these proteins and their isoforms can be viewed in QuickGO.
# Approved symbol Approved name UniProt ID 1 ABCA1 ATP binding cassette subfamily A member 1 O95477 2 ABCG1 ATP binding cassette subfamily G member 1 P45844 3 ADIPOQ adiponectin, C1Q and collagen domain containing Q15848 4 AGTR1 angiotensin II receptor type 1 P30556 5 APOB apolipoprotein B P04114 6 CD36 CD36 molecule P16671 7 CRP C-reactive protein P02741 8 CSF1 colony stimulating factor 1 P09603 9 CSF2 colony stimulating factor 2 P04141 10 ITGAV integrin subunit alpha V P06756 11 ITGB3 integrin subunit beta 3 P05106 12 LPL lipoprotein lipase P06858 13 MAPK9 mitogen-activated protein kinase 9 P45984 14 MPO myeloperoxidase P05164 15 MSR1 macrophage scavenger receptor 1 P21757 16 NFKB1 nuclear factor kappa B subunit 1 P19838 17 NFKBIA NFKB inhibitor alpha P25963 18 NR1H2 nuclear receptor subfamily 1 group H member 2 P55055 19 NR1H3 nuclear receptor subfamily 1 group H member 3 Q13133 20 PF4 platelet factor 4 P02776 21 PLA2G10 phospholipase A2 group X O15496 22 PLA2G2A phospholipase A2 group IIA P14555 23 PPARA peroxisome proliferator activated receptor alpha Q07869 24 PPARG peroxisome proliferator activated receptor gamma P37231 25 SOAT1 sterol O-acyltransferase 1 P35610 - Cardiac physiology
The goal of this BHF-funded project was to capture biological information about cardiac physiology. This project and its impact on data interpretation is described in Lovering et al. 2018. The list of priority proteins and microRNAs (listed below) were selected from a review of the literature. The annotations associated with these proteins and their isoforms and these microRNAs can be viewed in QuickGO.
# Approved symbol Approved name UniProt ID/RNA central ID 1 ACE angiotensin I converting enzyme P12821 2 ACE2 angiotensin I converting enzyme 2 Q9BYF1 3 AGT angiotensinogen P01019 4 AKAP6 A-kinase anchoring protein 6 Q13023 5 AKAP9 A-kinase anchoring protein 9 Q99996 6 ANK2 ankyrin 2 Q01484 7 ANK3 ankyrin 3 Q12955 8 ASPH aspartate beta-hydroxylase Q12797 9 ATP1A1 ATPase Na+/K+ transporting subunit alpha 1 P05023 10 ATP1A2 ATPase Na+/K+ transporting subunit alpha 2 P50993 11 ATP1A3 ATPase Na+/K+ transporting subunit alpha 3 P13637 12 ATP1B1 ATPase Na+/K+ transporting subunit beta 1 P05026 13 ATP2A2 ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 P16615 14 ATP2B4 ATPase plasma membrane Ca2+ transporting 4 P23634 15 CACNA1C calcium voltage-gated channel subunit alpha1 C Q13936 16 CACNA1D calcium voltage-gated channel subunit alpha1 D Q01668 17 CACNA1G calcium voltage-gated channel subunit alpha1 G O43497 18 CACNA2D1 calcium voltage-gated channel auxiliary subunit alpha2delta 1 P54289 19 CACNB2 calcium voltage-gated channel auxiliary subunit beta 2 Q08289 20 CALM1 calmodulin 1 P62158 21 CAMK2D calcium/calmodulin dependent protein kinase II delta Q13557 22 CASQ2 calsequestrin 2 O14958 23 CAV1 caveolin 1 Q03135 24 CAV2 caveolin 2 P51636 25 CAV3 caveolin 3 P56539 26 CLIC2 chloride intracellular channel 2 O15247 27 CNOT1 CCR4-NOT transcription complex subunit 1 A5YKK6 28 DLG1 discs large MAGUK scaffold protein 1 Q12959 29 DMD dystrophin P11532 30 DSG2 desmoglein 2 Q14126 31 FGF12 fibroblast growth factor 12 P61328 32 FGF13 fibroblast growth factor 13 Q92913 33 FKBP1B FK506 binding protein 1B P68106 34 FLNA filamin A P21333 35 FXYD1 FXYD domain containing ion transport regulator 1 O00168 36 GJA1 gap junction protein alpha 1 P17302 37 GJA5 gap junction protein alpha 5 P36382 38 GPD1L glycerol-3-phosphate dehydrogenase 1 like Q8N335 39 GSTM2 glutathione S-transferase mu 2 P28161 40 HCN4 hyperpolarization activated cyclic nucleotide gated potassium channel 4 Q9Y3Q4 41 HRC histidine rich calcium binding protein P23327 42 KCNA5 potassium voltage-gated channel subfamily A member 5 P22460 43 KCND3 potassium voltage-gated channel subfamily D member 3 Q9UK17 44 KCNE1 potassium voltage-gated channel subfamily E regulatory subunit 1 P15382 45 KCNE2 potassium voltage-gated channel subfamily E regulatory subunit 2 Q9Y6J6 46 KCNE3 potassium voltage-gated channel subfamily E regulatory subunit 3 Q9Y6H6 47 KCNE5 potassium voltage-gated channel subfamily E regulatory subunit 5 Q9UJ90 48 KCNH2 potassium voltage-gated channel subfamily H member 2 Q12809 49 KCNJ2 potassium voltage-gated channel subfamily J member 2 P63252 50 KCNJ5 potassium voltage-gated channel subfamily J member 5 P48544 51 KCNJ8 potassium voltage-gated channel subfamily J member 8 Q15842 52 KCNQ1 potassium voltage-gated channel subfamily Q member 1 P51787 53 LIG3 DNA ligase 3 P49916 54 LITAF lipopolysaccharide induced TNF factor Q99732 55 NDRG4 NDRG family member 4 Q9ULP0 56 NEDD4L neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase Q96PU5 57 NOS1 nitric oxide synthase 1 P29475 58 NOS1AP nitric oxide synthase 1 adaptor protein O75052 59 NPPA natriuretic peptide A P01160 60 NUP155 nucleoporin 155 O75694 61 PDE4B phosphodiesterase 4B Q07343 62 PDE4D phosphodiesterase 4D Q08499 63 PKP2 plakophilin 2 Q99959 64 PLN phospholamban P26678 65 PRKACA protein kinase cAMP-activated catalytic subunit alpha P17612 66 PTPN3 protein tyrosine phosphatase, non-receptor type 3 P26045 67 RANGRF RAN guanine nucleotide release factor Q9HD47 68 RNF207 ring finger protein 207 Q6ZRF8 69 RYR2 ryanodine receptor 2 Q92736 70 SCN10A sodium voltage-gated channel alpha subunit 10 Q9Y5Y9 71 SCN1B sodium voltage-gated channel beta subunit 1 Q07699 72 SCN2B sodium voltage-gated channel beta subunit 2 O60939 73 SCN3B sodium voltage-gated channel beta subunit 3 Q9NY72 74 SCN4B sodium voltage-gated channel beta subunit 4 Q8IWT1 75 SCN5A sodium voltage-gated channel alpha subunit 5 Q14524 76 SLC8A1 solute carrier family 8 member A1 P32418 77 SLC8B1 solute carrier family 8 member B1 Q6J4K2 78 SLC9A1 solute carrier family 9 member A1 P19634 79 SLMAP sarcolemma associated protein Q14BN4 80 SNTA1 syntrophin alpha 1 Q13424 81 SPTBN4 spectrin beta, non-erythrocytic 4 Q9H254 82 SRI sorcin P30626 83 TCAP titin-cap O15273 84 TRDN triadin Q13061 85 TRPM4 transient receptor potential cation channel subfamily M member 4 Q8TD43 86 YWHAE tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon P62258 87 YWHAH tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta Q04917 88 hsa-miR-1-3p miR-1 URS00001DC04F_9606 89 hsa-miR-26a-5p miR-26 URS000019B0F7_9606 90 hsa-miR-133a-3p miR-133a URS00004C9052_9606 91 hsa-miR-208a-3p miR-208a URS00000E5433_9606 92 hsa-miR-212-3p miR-212 URS00001D6BAE_9606 93 hsa-miR-328-3p miR-328 URS00005FDE70_9606 94 hsa-miR-499-5p miR-499 URS00000C7662_9606 - Insulin signaling
The goal of this BHF-funded project was to capture biological information about insulin signaling. The list of priority proteins (listed below) were selected from a review of the literature. The annotations associated with these proteins and their isoforms can be viewed in QuickGO.
# Approved symbol Approved name UniProt ID 1 AKT2 AKT serine/threonine kinase 2 P31751 2 APPL1 adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 Q9UKG1 3 CBL Cbl proto-oncogene P22681 4 GSK3A glycogen synthase kinase 3 alpha P49840 5 INS insulin P01308 6 INSR insulin receptor P06213 7 IRS1 insulin receptor substrate 1 P35568 8 IRS2 insulin receptor substrate 2 Q9Y4H2 9 PDE3B phosphodiesterase 3B Q13370 10 PDPK1 3-phosphoinositide dependent protein kinase 1 O15530 11 PRKCI protein kinase C iota P41743 12 PRKCZ protein kinase C zeta Q05513 13 RHOQ ras homolog family member Q P17081 14 SHC1 SHC adaptor protein 1 P29353 15 SORBS1 sorbin and SH3 domain containing 1 Q9BX66 - Atrial fibrillation - proteins
The goal of this project was to capture biological information about atrial fibrillation. The priority proteins (listed below) were selected from a review of the literature, for example Lozano-Velasco et al. (2020) lists the genes associated with atrial fibrillation, many of which overlap with the genes curated as part of the cardiac physiology project listed above.
# Approved symbol Approved name UniProt ID 1 ABCC9 ATP binding cassette subfamily C member 9 O60706 2 ANGPTL2 angiopoietin like 2 Q9UKU9 3 CACNA2D4 calcium voltage-gated channel auxiliary subunit alpha2delta 4 Q7Z3S7 4 CACNB2 calcium voltage-gated channel auxiliary subunit beta 2 Q08289 5 COL12A1 collagen type XII alpha 1 chain Q99715 6 COL21A1 collagen type XXI alpha 1 chain Q96P44 7 COL23A1 collagen type XXIII alpha 1 chain Q86Y22 8 COLQ collagen like tail subunit of asymmetric acetylcholinesterase Q9Y215 9 COMP cartilage oligomeric matrix protein P49747 10 GATA4 GATA binding protein 4 P43694 11 GATA5 GATA binding protein 5 Q9BWX5 12 GATA6 GATA binding protein 6 Q92908 13 GJA1 gap junction protein alpha 1 P17302 14 GJA5 gap junction protein alpha 5 P36382 15 GREM2 gremlin 2, DAN family BMP antagonist Q9H772 16 HCN4 hyperpolarization activated cyclic nucleotide gated potassium channel 4 Q9Y3Q4 17 KCNA5 potassium voltage-gated channel subfamily A member 5 P22460 18 KCND3 potassium voltage-gated channel subfamily D member 3 Q9UK17 19 KCNE1 potassium voltage-gated channel subfamily E regulatory subunit 1 P15382 20 KCNE2 potassium voltage-gated channel subfamily E regulatory subunit 2 Q9Y6J6 21 KCNE3 potassium voltage-gated channel subfamily E regulatory subunit 3 Q9Y6H6 22 KCNE4 potassium voltage-gated channel subfamily E regulatory subunit 4 Q8WWG9 23 KCNE5 potassium voltage-gated channel subfamily E regulatory subunit 5 Q9UJ90 24 KCNH2 potassium voltage-gated channel subfamily H member 2 Q12809 25 KCNJ2 potassium inwardly rectifying channel subfamily J member 2 P63252 26 KCNJ5 potassium inwardly rectifying channel subfamily J member 5 P48544 27 KCNJ8 potassium inwardly rectifying channel subfamily J member 8 Q15842 28 KCNN3 potassium calcium-activated channel subfamily N member 3 Q9UGI6 29 KCNQ1 potassium voltage-gated channel subfamily Q member 1 P51787 30 LMNA lamin A/C P02545 31 MMP3 matrix metallopeptidase 3 P08254 32 NKX2-5 NK2 homeobox 5 P52952 33 NKX2-6 NK2 homeobox 6 A6NCS4 34 NPPA natriuretic peptide A P01160 35 NUP155 nucleoporin 155 O75694 36 RyR2 ryanodine receptor 2 Q92736 37 SCN10A sodium voltage-gated channel alpha subunit 10 Q9Y5Y9 38 SCN1B sodium voltage-gated channel beta subunit 1 Q07699 39 SCN2B sodium voltage-gated channel beta subunit 2 O60939 40 SCN3B sodium voltage-gated channel beta subunit 3 Q9NY72 41 SCN4B sodium voltage-gated channel beta subunit 4 Q8IWT1 42 SCN5A sodium voltage-gated channel alpha subunit 5 Q14524 - Atrial fibrillation - microRNAs
The goal of this project was curate microRNAs that may impact on atrial fibrillation by regulating the levels of proteins associated with this disease. The priority microRNAs (listed below) that regulates proteins involved in electrial physiology were selected from a review of the literature, for example Lozano-Velasco et al. (2020) describes the miRNAs and lncRNAs networks controlling structural and electrical remodelling in atrial fibrillation.
# microRNA Target approved gene symbol 1 hsa-miR-1 HCN2 hsa-miR-1 HCN4 hsa-miR-1 KCNE1 hsa-miR-1 KCNB2 hsa-miR-1 KCNJ2 2 hsa-miR-106b RYR2 3 hsa-miR-133 HCN2 hsa-miR-133 HCN4 4 hsa-miR-192-5p SCN5A 5 hsa-miR-206 GJA1 6 hsa-miR-208 GJA5 7 hsa-miR-208b SERCA 8 hsa-miR-208b CACNA1C 9 hsa-miR-21 CACNA1C hsa-miR-21 CACNB2 10 hsa-miR-24 RYR2 hsa-miR-25 RYR2 11 hsa-miR-26 KCNJ2 12 hsa-miR-29 CACNA1C 13 hsa-miR-30d CACNA1C hsa-miR-30d KCNJ3 14 hsa-miR-328 CACNA1C hsa-miR-328 CACNB1 15 hsa-miR-384 CACNA1C 16 hsa-miR-499 KCNN3 hsa-miR-499 CACNB2
 Close
Close










