We manually curate proteins with an expected or known role in cardiovascular and neurological systems through the application of Gene Ontology (GO) terms and the capture of protein interaction data
We submit protein interaction data to the GO Consortium and to the IntAct resources, enabling the data to be downloaded from the PSICQUIC web server.
- Cardiovascular Gene Ontology protein annotation
- The British Heart Foundation (BHF) funded the creation of over 28,000 GO annotations which have facilitated the detailed and high-quality annotation of over 3,200 cardiovascular-relevant proteins, and there are many ways in with the impact of our curation on data analysis and annotation resources can be demonstrated. Over 41,000 protein annotations contributed by this project to the GO Consortium dataset are attributed to BHF-UCL following BHF grants SP/07/007/23671, 2007-2012; RG/13/5/30112, 2013-2018.
We initially prioritised 4,000 cardiovascular-relevant proteins:
- How the list was generated
- Browse the protein list
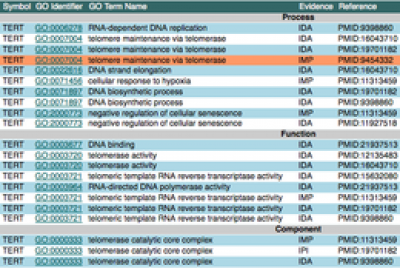
- Example of curated protein: human APOA4
Prioritised processes include:
- miRNA processing (Rachael)
- Telomerase and telomere-related proteins (Nancy and Jess)
- TGF-beta signaling pathway in heart development (Alexander Deng, MSc project student)
- Folic acid metabolism (Hadil Alrohaif, MSc project student)
- Hereditary hemochromatosis (Klaus Mitchell, MSc project student)
- Apoptosis (Ruth)
- Heart development (Varsha, Ruth)
- Insulin signaling (Ruth)
- Lipid metabolism (Ruth and Fiona Ratnaraj, MSc project student)
- Immunity (Evelyn)
- Notch pathway in heart development (Greg Rowe, MSc project student)
- Transcription factors associated with heart development (Varsha)
- Heart jogging (Varsha)
- BMP signalling pathway (Varsha)
- Inhibitory SMAD signalling pathway (Varsha)
- Growth factor regulated SMAD signalling pathway (Varsha)
- TGF-beta regulated SMAD signalling pathway (Varsha)
- Inositol 1,4,5-triphosphate-sensitive calcium-release channel pathway (Varsha)
- Growth hormone release pathway (Varsha)
- Reverse cholesterol transport (Ruth)
- Ryanodine-sensitive calcium-release channel activity (Varsha)
- Telomere maintenance (Ruth, Nancy)
- Transcriptional regulation by TCF7L2 (Ruth)
- Vitamin D metabolism (Varsha)
- Lipid trait risk associated genes (Ruth)
- CAFA2 project list (Anna, Ruth)
- Cardiac electrophysiology (Ruth)
- Alzheimer's disease-relevant Gene Ontology protein annotation
This initiative focused on the annotation of Alzheimer's disease-relevant proteins was funded by two Alzheimer's Research UK grants (ARUK-NSG2016-3, ARUK-NAS2017A-1) from January 2017 to date. By the end of 2019 the ARUK had funded the creation of over 5,000 GO annotations which have facilitated the detailed and high-quality annotation of over 650 Alzheimer's disease-relevant human proteins, and there are many ways in with the impact of our curation on data analysis and annotation resources can be demonstrated. Over 7,200 protein annotations contributed by this project to the GO Consortium dataset are attributed to ARUK-UCL.
Prioritised processes include:
- Amyloid-beta binding (Barbara)
- Tau biology (Barbara)
- Microglial activation (Barbara and Mila)
- Neuroinflammation
- Parkinson's disease-relevant Gene Ontology protein annotation
This initiative focused on the annotation of Parkinson's disease-relevant proteins and was funded by a 3-year Parkinson's UK grant (G-1307) from January 2014 to December 2016. Over 4,500 GO annotations to 800 Parkinson's disease-relevant human proteins have been created. Annotations contributed by this project to the GO Consortium dataset are attributed to ParkinsonsUK-UCL.
A high-priority set of 48 Parkinson's disease-relevant genes were selected based on genetic linkage or association studies.
Our longer Parkinson's-relevant priority list includes 329 gene products. These were chosen based on their interaction with high-priority gene products or because of their role in Parkinson's disease-related biological processes
browse the Parkinson's-relevant protein listGO annotations resulting from Parkinson's UK-funded projects
The following processes were prioritised, following discussions with experts in the field. These processes are considered to have important roles in Parkinson's disease.
- UCL-based Human Gene Nomenclature Committee
While based at UCL, HGNC provided Gene Ontology (GO) annotation of proteins during the process of assigning approved gene names and symbols, following funding by the Wellcome Trust, from 2004 - 2007. The project co-ordinator was Ruth Lovering and over 5000 GO annotations were created. Annotations contributed by this project to the GO Consortium dataset are attributed to HGNC-UCL.
- MSc student projects
We provide a variety of annotation projects for UCL undergraduate and graduate students. These have included:
- Describing the role of the TGF beta superfamily signalling pathway in embryonic cardiac valvular development using Gene Ontology annotations
- Functional annotation of proteins involved in lipid transport and metabolism
- Functional annotation of folate-related genes using the Gene Ontology in relation to pregnancy and fetal development
- Bioinformatics annotation of the proteins involved in Hemochromatosis
- Describing the role of the Notch signaling pathway in cardiac development using a bioinformatic approach
- Comprehensive Gene Ontology annotations on synaptogenesis and behavioural response associated with autistic spectrum disorder
 Close
Close

