There are many ways in which GO data is used to inform research decisions, from describing the roles of a gene product to analyses aimed at interpreting large-scale proteomic or transcriptomic data.
- Functional Gene Annotation papers demonstrating impact
- Lovering RC, et al. 2018 Improving Interpretation of Cardiac Phenotypes and Enhancing Discovery With Expanded Knowledge in the Gene Ontology
- Kramarz B, et al. 2018 Improving the Gene Ontology Resource to Facilitate More Informative Analysis and Interpretation of Alzheimer’s Disease Data
- Huntley RP, et al. 2018 Expanding the horizons of microRNA bioinformatics
- Denny P, et al. 2018 Exploring autophagy with Gene Ontology
- Foulger RE, et al. 2016 Using the Gene Ontology to Annotate Key Players in Parkinson's Disease
- Patel S, et al. 2015 Using Gene Ontology to describe the role of the neurexin-neuroligin-SHANK complex in human, mouse and rat and its relevance to autism
- Khodiyar VK, et al. 2013 From zebrafish heart jogging genes to mouse and human orthologs: using Gene Ontology to investigate mammalian heart development
- Alam-Faruque, et al. 2011 The impact of focused Gene Ontology curation of specific mammalian systems
- Khodiyar VK, et al. 2011 The representation of heart development in the gene ontology
- Lovering RC, et al. 2009 Improvements to cardiovascular gene ontology.
- TGF-beta signalling pathway in heart development (MSc project)
As a result of this curation project, we now have a improved the representation of the role of the TGFβ signalling pathway in heart development
Project
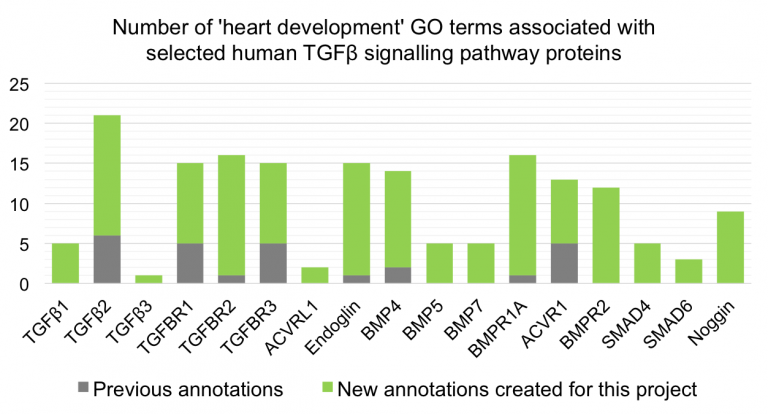
The GO annotation of the heart development role of 17 key proteins from the TGFβ signalling pathway.
Background
- In mammalian heart development, the endocardial cushions are the primitive structures which give rise to the cardiac valves and septa
- The TGFβ signalling pathway is known to be fundamentally involved in the formation of the endocardial cushions via the process of epithelial-to-mesenchymal transition
- A substantial body of primary literature describing the complex regulatory pathways of endocardial cushion development exists
- The usability of this data is limited as it is not available in bioinformatic databases for computational analysis
Impact
- At the start of this project, only 26 GO annotations pertaining to 'heart development' were present for the key human proteins of the TGFβ signalling pathway
- Nine of these proteins, including two of the three TGFβ proteins themselves, had 0 relevant annotations (see figure)
- At the end of the project, 146 GO annotations pertaining to 'heart development' were associated with these human proteins, an increase of more than 550%
- Re-analysis of a hypertension microarray dataset
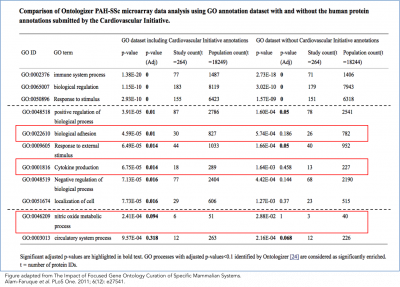
We have demonstrated an improved interpretation of the PAH-SSc dataset with the annotations supplied by the Cardiovascular GO Annotation Initiative (Alam-Faruque et al., 2011).
Project
Re-analysis of a microarray dataset of differentially regulated genes in peripheral blood mononuclear cells from patients with systemic scleroderma-related pulmonary arterial hypertension (PAH-SSc) compared to healthy controls.
Background
- In 2007 only 12 terms for heart development processes were available in GO
- The Cardiovascular GO Annotation Initiative was set up to improve GO for cardiovascular science
- We collaborated with ontology developers and heart experts to create new GO terms covering these complex processes
- We undertook manual literature-based annotation of cardiovascular-related proteins
- In order to to examine the impact of these efforts on analysis of high-throughput datasets, a cardiovascular-related microarray dataset was chosen for reanalysis
- We performed GO term enrichment analysis of a dataset published in 2008 (Grigoryev et al. Transl Res. 2008 Apr;151(4):197-207)
Impact
- Our hypothesis was: The British Heart Foundation funded GO annotations that we had created up until 2011 had positively impacted the interpretation of this dataset
- We performed GO term enrichment analysis using two datasets from 2011:
- i) the complete GO annotation dataset
- ii) the GO annotation dataset with the 13,000 Cardiovascular Initiative annotations removed
- Removing the Cardiovascular Initiative annotations decreased the significance of the majority of the enriched GO terms, and 6 GO terms were no longer significantly enriched.
- Several of these 6 GO terms are relevant to the PAH-SSc disease phenotype; e.g. 'cytokine production' and 'nitric oxide metabolic process' (see figure)
- Functional Gene Annotation Newsletters demonstrating impact
 Close
Close