MS data exchange
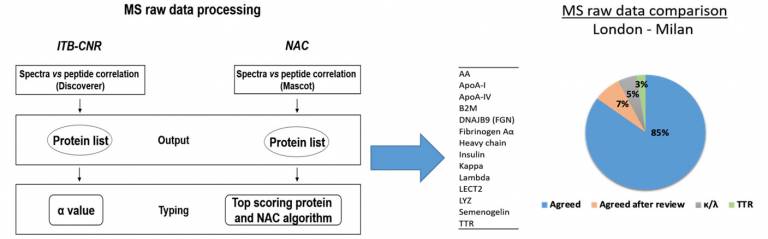
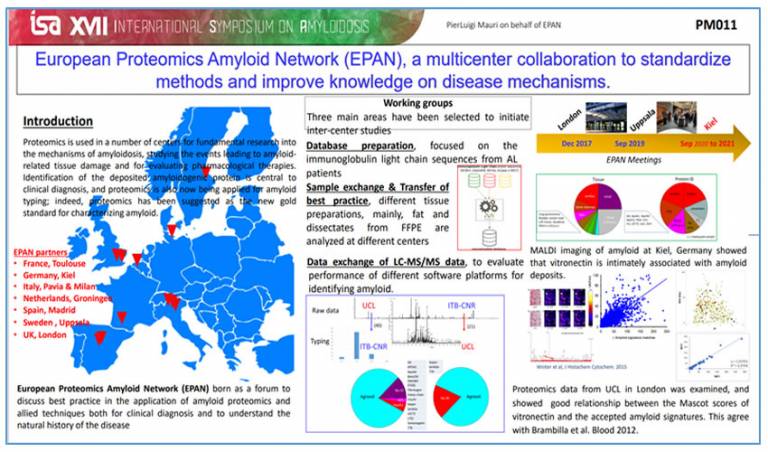
In the context of WG1, successful comparison has been obtained from the MS data exchange working group. MS data were shared between London (UCL-NAC) and Milan (ITB-CNR) facilities and the results showed very good reproducibility in terms of amyloid identification. An inter‐centre validation study was carried out comparing proteomics data obtained through different software platforms and bioinformatics tools at London (UCL-NAC) and Milan (ITB-CNR) centres. The work showed a high concordance (>92%) between the proteomics data (Canetti D et al, Molecules. 2021).
Sample Exchange - Work in progress
Database preparation
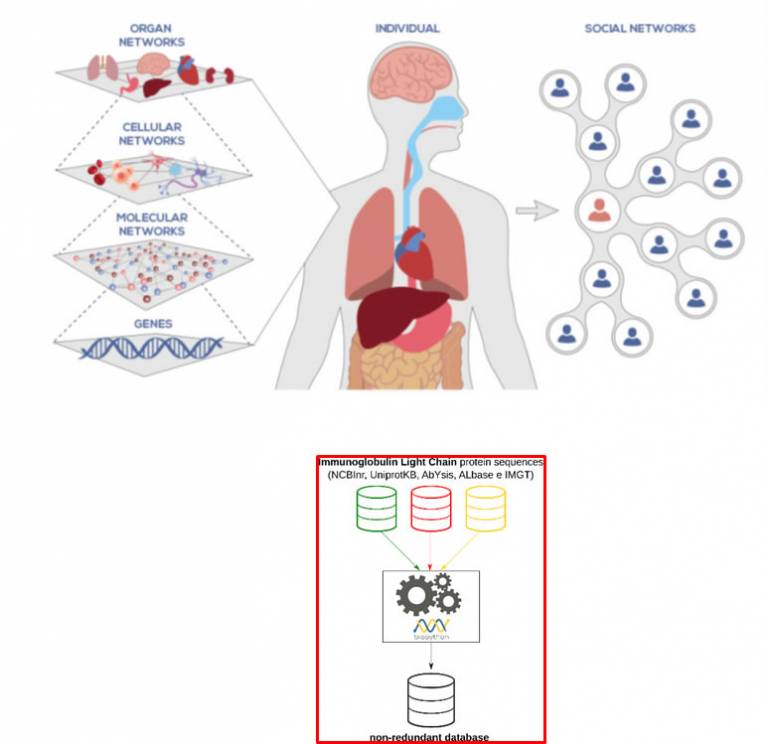
In the context of WG3 the activity concerns the reconstruction of a database containing unique immunoglobulin light chain from AL patients.
In order to reach this goal, we are developing a computational tool to automatically compare, classify, update and store the immunoglobulin light chain sequences available from the major public databases, including Uniprot, NCBI and ALbase. The resulting not redundant database will be useful for diagnostic and profiling purposes relying on mass spectrometry-based proteomic approaches.
Transfer of best practices
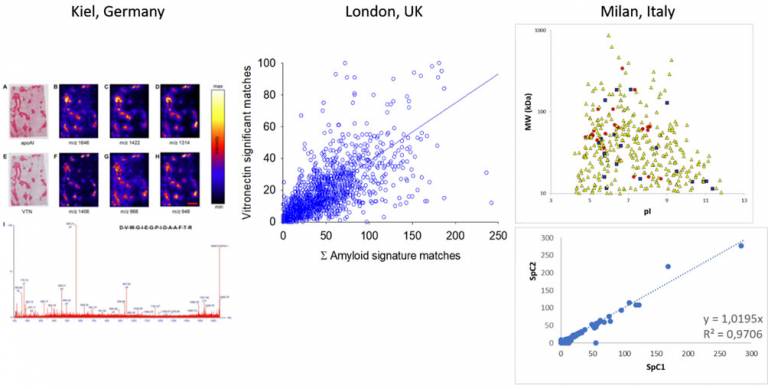
MALDI imaging of amyloid at Kiel showed that Vitronectin is intimately associated with amyloid deposits (Winter et al, J Histochem Cytochem. 2015). Proteomics data from UCL in London were examined, and showed good relationship between the Mascot scores of Vitronectin and the accepted amyloid signatures (ApoA-IV, ApoE and SAP). This data are also in agreement with the work performed from the ITB-CNR group (Brambilla et al, Blood. 2012).
Presentation to events



 Close
Close

