The Gene Ontology (GO) is the established standard for the functional annotation of gene products. GO is a controlled vocabulary used to classify the following functional attributes of gene products:
- Biological Process (BP) terms describe the biological processes a gene product is involved in
- Molecular Function (MF) terms describe the activities and functions a gene product executes
- Cellular Component (CC) terms describe the subcellular or extracellular localisations of a gene product
The GO annotations are added directly to the Gene Ontology Annotation (GOA) database, at the European Bioinformatics Institute.
- View the annotations for LRRK2, an example of a comprehensively annotated protein
We are focused on the:
Structure
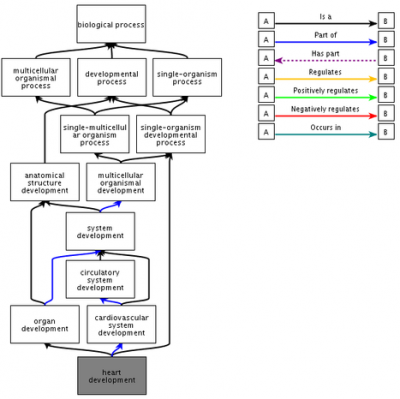
GO terms are structured in a directed graphs (see figure), where each term has relationships to broader 'parent' terms or more specific 'child' terms. This hierarchical structure produces a representation of biology that allows for a greater amount of flexibility in data analysis than would be afforded by a format based on simple list of terms. The figure shows the QuickGO representation of the ancestral chart (Parent Ontology) for the term 'heart development'.
 Close
Close

