Since January 2015 we have been using the Gene Ontology (GO) to describe the biological roles of microRNAs (miRNAs). To date we have provided all manual human RNA GO annotations.
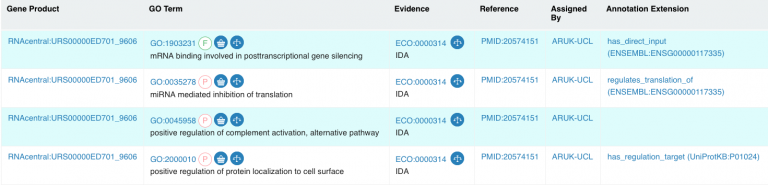
The RNA annotations are available in QuickGO, miRBase, NCBIGene, RNAcentral and AmiGO2, or in the regular GOA release files. In addition, our miRNA:target mRNA binding annotations are accessible for inclusion in network analyses from the PSICQUIC web service, in the EBI-GOA-miRNA file. Information about the data included in the PSICQUIC web service is also available.
Example: View the annotations associated with hsa-miR-133a-3p
miRNA curation guidelines
As part of this project we have drawn up miRNA curation guidelines (Huntley et al., 2016) in consultation with the Gene Ontology (GO) Consortium and experts in miRNA research, which cover how a curator should translate into GO annotations the common experimental assays used to investigate miRNA function, including how to annotate experimentally verified targets of miRNAs as well as the physiological effects that silencing has on the cell or organism. Note that GO Consortium guidelines have changed since the publication of the miRNA curation guidelines (Huntley et al., 2016). For more specific details see the Molecular interaction data set for miRNAs and their targets section below
Any queries about the miRNA project should be directed to r.lovering@ucl.ac.uk.
- Molecular interaction dataset for miRNAs and their targets
- The miRNA:mRNA interaction file is available: https://www.ebi.ac.uk/QuickGO/psicquic-rna/webservices/current/search/interactor/* and via the PSCIQUIC web service. The data set is in PSI-MITAB 2.7 format, which is described at https://psicquic.github.io/MITAB27Format.html.This file contains the miRNA (RNAcentral ID) and the mRNA (UniProt ID) that it has been experimentally shown to bind and/or regulate.In this file the interactions are indicated as either:
- 'physical association', meaning the miRNA has been shown experimentally to bind and regulate the mRNA (usually a luciferase assay)
- 'association', meaning the miRNA has been shown experimentally to regulate expression of the mRNA (e.g. by western blot or RT-PCR), but there was no experiment to demonstrate binding
- EBI-GOA-miRNA curation details
Huntley, et al (2018) provides a description of how the PSI-MITAB format interactions are derived from the GO annotations:
A molecular interaction dataset (“EBI-GOA-miRNA”) was created in PSI-MI format and made available on the PSICQUIC web service, to enable computational access to miRNA interactions with their experimentally validated targets. The source of the information in this data set is GO annotations that we have created containing experimentally verified miRNA:target interaction data. GO annotations used for this purpose conform to the following criteria; the “Database Object ID” field of the GO annotation file must be a miRNA, specified by an RNAcentral ID, AND the “Annotation Extension” field must contain an mRNA target, specified by a UniProt ID (del-Toro et al. 2013; Huntley et al. 2014). For the PSICQUIC specification, those interactions described with the Molecular Function GO term mRNA binding involved in post-transcriptional gene silencing (GO:1903231) are assigned interaction type “physical association,” indicating direct binding of the miRNA to the mRNA target. Interactions described only with one of the following GO Biological Process terms: gene silencing by miRNA (GO:0035195); miRNA mediated inhibition of translation (GO:0035278); mRNA cleavage involved in gene silencing by miRNA (GO:0035279); deadenylation involved in gene silencing by miRNA (GO:0098806), but without the Molecular Function term above are assigned the interaction type “association,” indicating that the evidence demonstrated miRNA regulation of the target only (see “Data set availability” section below for the file format information and access to this data set).
miRNA annotation projects undertaken
- Alzheimer's Research UK grant ARUK-NSG2018A-003
The ARUK funded the creation of over 1,000 GO annotations which have facilitated the detailed and high-quality annotation of 160 Alzheimer's-relevant microRNAs. A variety of focused projects were undertaken including the curation of:
- microRNAs that regulate microglial proteins
- The goal of this project was to annotate the microRNAs (miRNAs) that regulated the expression of the proteins with a role in microglial processes. Over 100 human miRNAs were annotated by Rachael Huntley and Barbara Kramarz, including hsa-miR-520b-3p, which we have annotated as regulating the expression of C3 and as positive regulation of complement activation, alternative pathway. In addition, 19 papers describing the experimentally verified mRNA targets of IL6 were curated, creating 90 annotations, and annotations describing 'neuron projection development' and 'gliogenesis' were contributed, yielding 41 annotations from 19 papers and 57 annotations from 13 papers, respectively.
- Alzheimer's Disease microRNA biomarkers
- Swarbrick S, et al., 2019, decribed a panel of 10 microRNAs suggested to be disregulated early in Alzheimer's disease. Barbara Kramarz curated 14 papers describing the microRNA target interactions for these microRNAs, when available, creating 54 annotations for 10 human microRNAs, including the potential biomarkers hsa-miR-146a-5p, hsa-miR-107, hsa-miR-26b-5p and hsa-miR-210-3p, and 9 proteins.
- microRNAs that regulate microglial proteins
- British Heart Foundation (BHF) grant RG/13/5/30112
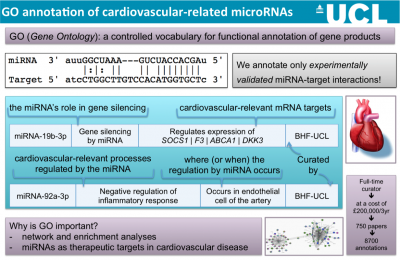
The BHF funded the creation of over 3,600 GO annotations which have facilitated the detailed and high-quality annotation of almost 400 cardiovascular-relevant microRNAs. A variety of focused projects were undertaken including the curation of:
- BHF-funded research papers
- From 2014 to 2018, Rachael Huntley curated all BHF funded research papers describing the role of microRNAs in cardiovascular-relevant processes.
- Regulation of cardiac physiology
- Rachael Huntley curated 7 microRNAs, as these had been identified as modulating cardiac excitability; literature available for these miRNAs was curated up to March 2016.
- Angiogenesis
- Over a period of 3 months Vanessa Acquaah curated the role of microRNAs in angiogenesis. The two major cell types are involved in this process: endothelial and smooth muscle cells (SMC). During this project Vanessa annotated 114 papers that demonstrate the roles that miRNAs play in regulating endothelial and SMC cell proliferation, migration and endothelial tube formation. These papers were identified by searches in pubmed using the key terms “mirnas in angiogenesis”. GO annotations associated with a selection of these curated papers are available in QuickGO.
- Pulmonary artery remodelling
- Pulmonary hypertension is caused by impaired pulmonary arterial growth and remodelling. Vanessa Acquaah annotated 24 papers that describe the role of miRNAs in vascular smooth muscle cell pulmonary artery remodelling, creating 167 annotations. These papers were identified by searches in pubmed using the key words “miRNAs and pulmonary smooth muscle cells”.
- BHF-funded research papers
- Familial hypercholesterolemia, MSc project
Hao Chen, MSc project 2017. Familial hypercholesterolemia (FH) is a common inherited lipid disorder that can cause cardiovascular diseases (CVD) if untreated. Treatment on FH depend on management of plasma low-density-lipoprotein (LDL) cholesterol. High level of plasma LDL cholesterol can induce atherosclerosis and plaque buildup in the walls of the arteries. There is evidence that suggest miRNAs’ regulation of LDL-C is due to their regulation of FH-associated genes, such as LDLR, PCSK9, APOB, LDLRAP1, ABCA1 and ABCG1. This project created 290 GO annotations, of which 22 were transferred from mouse miRNAs to human miRNAs using the ISS evidence code. The prioritised mRNA targets are listed below:
#
HGNC Symbol
HGNC Name
UniProt ID
1
ABCA1
ATP binding cassette subfamily A member 1
2
ABCG1
ATP binding cassette subfamily G member 1
3
APOB
apolipoprotein B
4
LDLR
low density lipoprotein receptor
5
LDLRAP1
low density lipoprotein receptor adaptor protein 1
6
PCSK9
proprotein convertase subtilisin/kexin type 9
- Angiogenesis, IBSc project
Marios Makris, IBSc project 2019. Angiogenesis is the formation of blood vessels from pre-existing vasculature, involving endothelial cell migration, proliferation, survival, vascular tube formation and regulation of angiogenic growth factors. A total of 237 GO annotations were created to associate 40 distinct microRNAs to 11 pro-angiogenic growth factors. The downstream angiogenesis-relevant effects of regulating these factors were also captured. The prioritised mRNA targets are listed below:
No.
Approved gene name
Approved Gene Symbol
1
Vascular endothelial growth factor A
VEGFA
2
Kinase insert domain receptor*
KDR*
3
Fibroblast growth factor 2
FGF2
4
Fibroblast growth factor receptor 1
FGFR1
5
Fibroblast growth factor receptor 2
FGFR2
- Aortic aneurysm, MSc project
Zara Umrao, MSc project 2015. Aortic aneurysm (AA) is a cardiovascular disease characterized by swelling of the aorta, the main vessel that runs from the heart to the abdomen. Recently, research has revealed the role of microRNAs (miRNA) in some diseases, including AA, which may enable a better understanding of the disease. The role of one such family of miRNAs, miRNA29, was investigated by creating annotations from existing literature. Forty papers were curated, from which 497 GO annotations were made. The annotations spanned a range of species, including human, mouse, rat, zebrafish and dog. The role of miRNA29 in regulating essential extracellular matrix proteins, such as elastin, collagen, fibrillin and matrix metalloproteinases was captured. In addition, the GO annotations also described the role of miRNA29s in activities, such as aorta morphogenesis, cell migration, cell proliferation, DNA methylation and epithelium regeneration.
- Early heart development, MSc project
Vanessa Acquaah, MSc project 2016. Recent evidence has shown that miRNAs, in particular microRNA-1 (miR-1) are found in abundance within cardiac tissues and may be particularly important in early heart development. A total of 313 annotations were made through the use of experimental evidence from 49 papers using GOC curation guidelines. Annotations covered the role of miR-1 as well as other miRNAs in cardiomyocyte proliferation and cardiomyocyte differentiation. Additionally, development of cardiac neural crest cells through processes such as endothelial to mesenchymal transition (EMT) and differentiation of epithelial cells are processes regulated by miRNAs; these roles have been captured as GO annotations.
- Regulation of amyloid-beta 'good' receptors, MSc project
Shirin Saverimuttu, MSc project 2018. Alzheimer’s disease (AD), the most common form of dementia, is characterised by amyloid beta (Aβ) accumulation in extracellular plaques. Moreover, Aβ accumulation has been hypothesised as a primary cause of the disease, with a dysregulation to the proteins involved in Aβ clearance thought to be critical. Interestingly, hundreds of microRNAs have been found to be altered in brain tissue affected by AD and several have been experimentally proven to regulate the expression of Aβ clearance proteins. This MSc project led to a total of 258 GO annotations being created. This allowed for knowledge describing how microRNAs regulate Aβ clearance proteins to be captured, as well as the capturing the downstream effects of these microRNAs. The prioritised mRNA targets are listed below:
#
HGNC Symbol
HGNC Name
UniProt ID
1
APOE
Apolipoprotein E
2
CHRNA7*
Cholinergic receptor nicotinic alpha 7 subunit
3
CLU
Clusterin
4
FPR2
Formyl peptide receptor 2
5
HSPG2
Heparan sulfate proteoglycan 2
6
ITGA2*
Integrin subunit alpha 2
7
ITGAM*
Integrin subunit alpha M
8
LRP1
LDL receptor related protein 1
9
LRP2
LDL receptor related protein 2
10
LDLR
Low density lipoprotein receptor
11
MARCO*
Macrophage receptor with collagenous structure
12
MSR1*
Macrophage scavenger receptor 1
13
PICALM*
Phosphatidylinositol binding clatherin assembly protein
14
PRNP
Prion protein
15
TLR2
Toll like receptor 2
16
TLR4
Toll like receptor 4
* there was no experimental evidence to support annotation of microRNAs regulating these proteins in July 2019
- Regulation of interleukins
Shirin Saverimuttu is responsible for this project, the goal of which is to annotate microRNAs (miRNAs) that regulate the expression of interleukins with a role in Alzheimer’s-relevant processes. In order to achieve this goal, it was necessary to generating a priority list of interleukins (below) and then to identify their regulatory miRNAs. The miRNAs are identified on a target-by-target basis, firstly using mirTarBase and then with specific searches in PubMed. This project is funded from Functional Gene Annotation group’s discretionary funds. The prioritised mRNA targets are listed below:
#
HGNC Symbol
HGNC Name
UniProt ID
1
IL1A
Interleukin 1 alpha
P01583
2
IL1B
Interleukin 1 beta
P01584
3
IL1F10
Interleukin 1 family member 10
Q8WWZ1
4
IL2
Interleukin 2
P60568
5
IL3
Interleukin 3
P08700
6
IL4
Interleukin 4
P05112
7
IL5
Interleukin 5
P05113
8
IL6
Interleukin 6
P05231
9
IL7
Interleukin 7
P13232
10
CXCL8
C-X-C motif chemokine ligand 8 (alias: interleukin 8)
P10145
11
IL9
Interleukin 9
P15248
12
IL10
Interleukin 10
P22301
13
IL11
Interleukin 11
P20809
14
IL12A
Interleukin 12A
P29459
15
IL12B
Interleukin 12B
P29460
16
IL13
Interleukin 13
P35225
17
IL15
Interleukin 15
P40933
18
IL16
Interleukin 16
Q14005
19
IL17A
Interleukin 17A
Q16552
20
IL17B
Interleukin 17B
Q9UHF5
21
IL17C
Interleukin 17C
Q9P0M4
22
IL17D
Interleukin 17D
Q8TAD2
23
IL17F
Interleukin 17F
Q96PD4
24
IL18
Interleukin 18
Q14116
25
IL19
Interleukin 19
Q9UHD0
26
IL20
Interleukin 20
Q9NYY1
27
IL21
Interleukin 21
Q9HBE4
28
IL22
Interleukin 22
Q9GZX6
29
IL23A
Interleukin 23 alpha
Q9NPF7
30
IL24
Interleukin 24
Q13007
31
IL25
Interleukin 25
P01589
32
IL26
Interleukin 26
Q9NPH9
33
IL27
Interleukin 27
Q8NEV9
34
IL31
Interleukin 31
Q6EBC2
35
IL32
Interleukin 32
P24001
36
IL33
Interleukin 33
O95760
37
IL34
Interleukin 34
Q6ZMJ4
38
IL36A
Interleukin 36 alpha
Q9UHA7
39
IL36B
Interleukin 36 beta
Q9NZH7
40
IL36G
Interleukin 36 gamma
Q9NZH8
41
IL37
Interleukin 37
Q9NZH6
- Endothelial connections at the Blood Brain Barrier
Katherine Thurlow, MSc project 2019. Evidence for cerebrovascular mechanisms, particularly increased breakdown and dysfunction of the blood-brain barrier (BBB), as key contributors to Alzheimer’s Disease (AD), the most common form of dementia, has grown in recent years,. The BBB is key in regulating influx and efflux of substances to the brain and as such maintains normal neuronal and cognitive function. Alterations in BBB constituent proteins could alter its ability to selectively transport molecules and result in a change in permeability to potentially toxic substances. The list of priority mRNA targets for this project was identified from Figure 2 of Sweeney et al., 2019. This MSc project led to a total of 253 GO annotations being created. This allowed for knowledge describing the role of microRNAs at the BBB to be captured. The prioritised mRNA targets are listed below:
# UniProt ID
HGNC Symbol
HGNC Name
1 P60709
ACTB
actin beta
2
P63261
ACTG1
actin gamma 1
3
P33151
CDH5
cadherin 5
4
O95832
CLDN1
claudin 1
5
P56749
CLDN12
claudin 12
6
O15551
CLDN3
claudin 3
7
O00501
CLDN5
claudin 5
8
P11532
DMD
dystrophin
9
Q96AP7
ESAM
endothelial cell adhesion molecule
10
Q9Y624
F11R
F11 receptor
11
P17302
GJA1
gap junction protein alpha 1
12
O95452
GJB6
gap junction protein beta 6
13
P05556
ITGB1
Integrin subunit beta 1
14
P57087
JAM2
junctional adhesion molecule 2
15
Q9BX67
JAM3
junctional adhesion molecule 3
16
P24043
LAMA2
Laminin subunit alpha-2
17
P11047
LAMC1
Laminin subunit gamma-1
18
Q86X29
LSR
lipolysis stimulated lipoprotein receptor
19
Q16625
OCLN
occludin
20
P16284
PECAM1
platelet and endothelial cell adhesion molecule 1
21
Q07157
TJP1
tight junction protein 1
22
Q9UDY2
TJP2
tight junction protein 2
23
O95049
TJP3
tight junction protein 3
24
P18206
VCL
vinculin
- Regulation of key proteins involved in APP processing and Aβ formation
Sandra De Miranda Pinheiro, MSc project 2019. Accumulation of amyloid-beta (Aβ) and tau proteins in the brain is the hallmark of Alzheimer’s disease and lead to neurodegeneration. An imbalance between the production and the clearance of Aβ from the brain is thought to drive the development of the disease. Aβ derives from a larger protein, the amyloid precursor protein (APP), which is processed by different secretases in a sequential way. In the amyloidogenic pathway, β-secretase cleaves APP at the β-site and cleavage of the remaining APP fragment by γ-secretase results in the production of Aβ peptides. Increased APP expression has been associated with an increased risk of developing Alzheimer’s disease. This MSc project led to a total of 254 GO annotations being created. This allowed for knowledge describing how microRNAs regulate the processing of APP and the formation of Aβ. The prioritised mRNA targets are listed below:
#
HGNC Gene Symbol
HGNC Approved Name
Protein Group
1
APP
amyloid beta precursor protein
2
ADAM9
ADAM metallopeptidase domain 9
Alpha-secretase
3
ADAM10
ADAM metallopeptidase domain 10
Alpha-secretase
4
ADAM17
ADAM metallopeptidase domain 17
Alpha-secretase
5
ADAM19
ADAM metallopeptidase domain 19
Alpha-secretase
6
APH1A
aph-1 homolog A
Gamma-secretase subunit
7
APH1B
aph-1 homolog B
Gamma-secretase subunit
8
BACE1
beta-secretase 1
Beta-Secretase 1
9
NCSTN
nicastrin
Gamma-secretase
10
PSEN1
presenilin 1
Gamma-secretase
11
PSEN2
presenilin 2
Gamma-secretase
12
PSENEN
presenilin enhancer
Gamma-secretase subunit
- Regulation of Aβ-binding 'bad' receptors
Diana Luna Buitrago, MSc project 2020. Amyloid-beta (Aβ) accumulation within the brain and abnormal processing of Aβ by various Aβ-binding receptors are known to be characteristics of Alzheimer's disease pathology. Through previous studies, Aβ-binding receptors have been identified to be “good” or “bad”. Here, “bad” receptors are those that bind to Aβ to promote downstream neurotoxic processes associated with AD. MicroRNAs may contribute to AD by acting as regulators of Aβ-binding receptor expression, as microRNAs that alter “bad” Aβ-binding receptor levels and may influence the neurotoxicity of Aβ. The table below lists the microRNAs curated to regulate the expression of known “bad” Aβ-binding receptors during this project:
MicroRNAs identified to regulate key proteins of interest
AMPA receptor subunits
Ephrin receptors
INSR
GRIA1*
GRIA2*
GRIA3*
GRIA4*
EPHA4
EPHB2
hsa-miR-124-3p
hsa-miR-124-3p
hsa-miR-181b-5p
hsa-miR-181a-5p
hsa-miR-124-3p
hsa-miR-28-5p
hsa-miR-10a-5p
hsa-miR-519d-3p
hsa-miR-335-5p
hsa-miR-145-5p
hsa-miR-185-5p
hsa-miR-520d-3p
hsa-miR-204-5p
hsa-miR-195-5p
hsa-miR-7-5p
hsa-miR-1271-5p
hsa-miR-15b-5p
Frizzled receptors
AGER
NGFR
FZD5
FZD4
hsa-miR-29c-3p
hsa-miR-515-5p
hsa-miR-224-5p
hsa-miR-493-3p
hsa-miR-199a-5p
hsa-miR-29c-3p
hsa-miR-185-5p
hsa-miR-127-5p
hsa-miR-98-5p
hsa-miR-296-5p
- Regulation of tau-associated processes
Miao Long, MSc project 2020. Alzheimer's disease (AD) is characterised neuropathologically as the presence of intraneuronal neurofibrillary lesions expressing the Tau protein. One of the characteristic pathologies of sporadic AD patients, is hyperphosphorylation of Tau and neurofibrillary tangles. Remarkably, hundreds of microRNAs have been shown to be changed in AD-affected brain regions, with some of them being experimentally demonstrated to regulate tau-related activities. This project focused on capturing the role of microRNAs in regulation of proteins involved in Tau hyperphosphorylation. The mRNA targets identified in this project are listed below:
# UniProt ID HGNC Symbol HGNC Name 1 P35222 CTNNB1 catenin beta 1 2 O94907 DKK1 dickkopf WNT signaling pathway inhibitor 1 3 P49841 GSK3B glycogen synthase kinase 3 beta 4 P10636 MAPT microtubule associated protein tau 5 Q9P0L2 MARK1 microtubule affinity regulating kinase 1 6 P27448 MARK3 microtubule affinity regulating kinase 3 7 Q96L34 MARK4 microtubule affinity regulating kinase 4 8 Q969G9 NKD1 NKD inhibitor of WNT signaling pathway 1 9 P67775 PPP2CA protein phosphatase 2 catalytic subunit alpha 10 P62714 PPP2CB protein phosphatase 2 catalytic subunit beta 11 Q13362 PPP2R5C protein phosphatase 2 regulatory subunit B'gamma 12 P10644 PRKAR1A protein kinase cAMP-dependent type I regulatory subunit alpha 13 Q04206 RELA RELA proto-oncogene, NF-kB subunit 14 Q13464 ROCK1 Rho associated coiled-coil containing protein kinase 1 15 P01375 TNF tumor necrosis factor 16 Q5TCY1 TTBK1 tau tubulin kinase 1
 Close
Close

