Ontology development is achieved both by the creation of individual terms across GO as they are required for curation, and also by the larger-scale revision of a focussed node of the ontology.
The UCL team has been involved in several ontology development projects, as detailed below:
| Topic | Project start date | Details |
| Transcription | January 2017 | Ruth is a member of the eCOST funded GREEKC project and is contributing to the improvements to this part of the ontology |
| Protein complexes | December 2016 | We are creating GO terms for protein complexes relevant to our curation focus. In addition, we are suggesting corresponding complexes in the IntAct Complex Portal so we can attach annotations to the complexes themselves. |
| Telomere maintenance | December 2016 | Nancy revised and requested telomere and telomerase-related terms in GO. This followed a result of a collaboration with Professor Elizabeth Blackburn, and was part of the BHF-funded annotation effort of heart disease-related processes. |
| Mitophagy and Autophagy | April 2015 | Paul worked with autophagy experts and curators from other databases to improve the representation of autophagy (and specifically mitophagy) in GO. This was part of annotation of Parkinson's disease-related processes. |
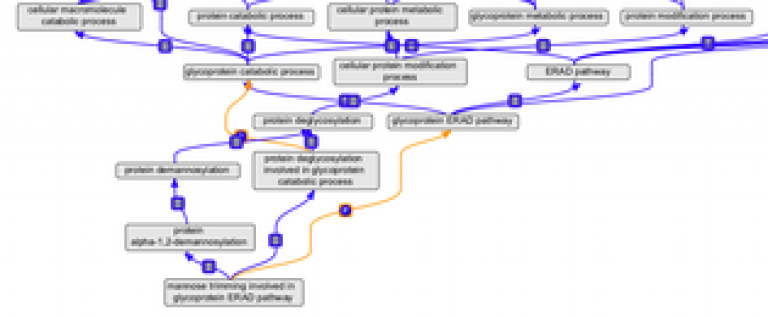
| Response to endoplasmic reticulum stress | October 2014 | Rebecca revised the ER stress response node of GO, to facilitate curation of Parkinson's disease-related processes. |
| Response to oxidative stress | June 2014 | Rebecca revised the oxidative stress response node of GO, to facilitate curation of Parkinson's disease-related processes. |
| Apoptosis | February 2011 | Ruth worked on the apoptosis ontology and attended a 1 day apoptosis workshop in June 2011 to refine the representation of apoptosis within the Gene Ontology. In May 2013 there was a GO Consortium co-ordinated reannotation of this domain. |
| Cardiac conduction | January 2010 | Ruth organised a cardiac conduction workshop in November 2011 to develop the ontology in this area. This led to the creation of over 120 new GO terms describing this domain of the ontology |
| Signaling | March 2009 | Ruth worked with GO editors, GO annotators and signaling experts to refine the representation of signaling with GO. This included helping organise a two-day signaling workshop in February 2011. |
| Heart development | January 2009 | Varsha organised a 1.5 day heart development workshop in September 2009 with cardiac development experts, GO editors and GO annotators from several model organism databases, to develop the ontology in this area. This project led to the creation of 285 new GO terms to describe heart development. The new ontology, presented in Khodiyar et al., 2011, describes heart morphogenesis, differentiation of specific cardiac cell types, and the involvement of signaling pathways in heart development. Following on from this successful workshop, Varsha led a Reference Genome Project to annotate transcription factors involved in heart development. |
| Cardiovascular lipoprotein metabolism | November 2007 | Ruth worked with cardiovascular lipoprotein experts to develop GO terms for describing biological characteristics of lipoproteins across the biological process, molecular function and cellular component nodes. |
Requests from the BHF-funded project have so far led to the creation of over 2000 GO terms, the Parkinson's UK-funded project led to the creation of over 450 GO terms, and the Alzheimer's Research UK-funded project has created or revised over 100 GO terms.
 Close
Close

