HDR UK National Text Analytics Workshop 12 March 2021
2 April 2021
Governance, tools and approaches for using text analytics and natural language processing for research

Introduction
Information stored within electronic health records (EHR) that is recorded in written form – sometimes referred to as unstructured text – is difficult to use in research because special computerised tools are needed to process the text and complex governance arrangements make such data in the NHS hard to access. Better use of unstructured text contained within EHR for research would have a number of benefits for patients including more streamlined matching of patients to clinical trials, better understanding of risk and outcome for diseases, and identification of drug-repurposing opportunities, among many others.
The HDR UK National Text Analytics Project, funded by Health Data Research UK (HDR UK) and led by Prof Richard Dobson and Dr Angus Roberts, is helping to build the UK's natural language processing (NLP) community for healthcare by making available shared tools, methods and datasets across the NHS, creating richer, more useful clinical information to improve healthcare.
On 12 March 2021, more than 85 people from across the UK text analytics community came together to discuss challenges of accessing unstructured text for research, new opportunities, and see how existing text analytics and NLP data extraction tools could be used in their research.
Summary of Sessions
ACCESSING UNSTRUCTURED DATA FOR RESEARCH (Chairs: Prof Rob Stewart, Professor of Psychiatric Epidemiology & Clinical Informatics, King’s College London and Dr Will Whiteley, Scottish Senior Clinical Fellow, University of Edinburgh)
Speakers:
• Dr Nathan Lea, Information Governance Manager for Research, UCLH NHS Foundation Trust and the NIHR Biomedical Research Centre Clinical Research Informatics Unit
• Jackie Caldwell, Information Consultant, Public Health Scotland
• Professor Rob Stewart, Professor of Psychiatric Epidemiology & Clinical Informatics, King’s College London
• Dr Dan Schofield, Data Scientist, NHSX
• Dr Elizabeth Ford, Senior Lecturer in Primary Care Research, University of Sussex
The text analytics and NLP community currently has few options available to them in terms of open and accessible datasets for use for research, and complex governance and ethics requirements remains a barrier. This session featured talks from information governance experts, data custodians and researchers with expertise in using free text to highlight key challenges to use of such data for research.
The data’s identifiable - that doesn’t mean you can’t use it. Although NLP methods are becoming ever more powerful, the problem is that our raw data processing is biased towards use of structured data . While unstructured text is never truly anonymous because records contain so much detail, it can still be used for research under the correct conditions. Removal of too much detail can however reduce the nuance in the text and this is therefore a hard balance to achieve. Researchers must ensure data are processed accurately, securely, accountably and transparently.
Generic governance models and a framework for use of free text. Two examples of generic governance models for research using free text were presented. Generation Scotland is a population and family-based study of genetic influences on disease and wellbeing including a wide range of health and lifestyle data. Free text (prescriptions (dosage), GP records (notes) and scan reports are made available for research under a model which encompasses the “Five Safes”: Safe People, Safe Data, Safe Projects, Safe Settings and Safe Output. Challenges include handling the volume (>2PB) and technical and computing challenges of managing such large datasets, and ensuring the de-identification of algorithms is maintained and remains accurate over time. Generation Scotland has generic ethics approval for research undertaken in their data safe haven. CRIS (Clinical Record Interactive Search) has enabled mental health research using EHR at South London and Maudsley NHS Trust through a governance framework that ensures patient anonymity and places service users at its core. The CRIS security model was developed and managed by a stakeholder / patient-led oversight committee.
NHS language corpus discovery project (NHSX) The NHS Language Corpus Discovery project is using open tools from the internet (e.g. from NHS.UK, NHS Data Dictionary) to understand what metadata or enrichment is useful and how best to share the outputs (can we, should we, how best to share?). The Corpus seeks to be open, representative, extensible (collect a dataset that has a wide coverage as well as a large number of examples over time) and useful (adds to the currently available resources constructively). Next steps will be to add datasets containing text from clinical settings and collect examples of synthetic text or successful de-identification that have already been approved, and add examples of datasets which sit closer to patient-centred interactions with the NHS. NHSX are keen to hear from anyone who has ideas around which data sources to add or feed into enriching their user stories.
Public engagement around healthcare text mining Public opinion defines what, as researchers, we can do with health data. Public involvement activities have greatly informed our knowledge around public attitudes to the use of their free text data for research and it has been found that people are generally willing to share their EHR data for the common good and people tend to change their minds as they become more informed about the area and the aims of what people are trying to do with text. Despite this work, there remains a gap in knowledge around public opinion that we need to fill. Addressing this gap is a crucial step towards informing and influencing data access policies. TexGov, led by Prof Kerina Jones in Swansea, is developing data governance toolkits and standards for use of clinical free text data in research. This work highlights that use of free text is not fundamentally different from use of structured data for research and includes recommendations for a donated bank of text for use by the text analytics community.
Crucial next steps involve development of a plan for how the public can be involved in free text research as collaborators and research team members, and how to include public members on decision making panels, including what information they need about our research.
DEMONSTRATION OF EXISTING TEXT ANALYTICS AND NLP TOOLS AND USE
IN RESEARCH TO IMPROVE CARE (Chairs: Professor Richard Dobson, Professor in Medical & Bio-Informatics, UCL Institute of Health Informatics and King's College London and Dr Angus Roberts, Senior Lecturer in Health Informatics, King’s College London)
Speakers:
• Dr James Teo, Neurologist & Joint Clinical Director of Data Science and AI, King's College Hospital and Guys & St Thomas Hospital
• Tom Searle, King’s College London
• Professor Nigel Collier, Professor of Natural Language Processing, University of Cambridge
• Andreas Grivas, Research Associate/PhD student, University of Edinburgh
• Dr Hang Dong, Research Fellow, Centre for Medical Informatics, Usher Institute, University of Edinburgh
• Dr Luke Slater, Research Fellow, University of Birmingham
• Dr Arlene Casey, Research Fellow, University of Edinburgh
• Jyoti Sanyal, NLP Lead, Maudsley Biomedical Research Centre, London
This session featured demonstrations of a number of open tools that have been developed across UK sites and use cases, including:
Tools
MedCAT (Medical Concept Annotation Toolkit) (King’s College London): deep Learning Neural Network learning to read medical text
CogStack (Maudsley Biomedical Research Centre): an application framework for information retrieval and extraction with uses in NLP, enterprise search, alerting, cohort selection and research
CRIS (Clinical Record Interactive Search) (Maudsley Biomedical Research Centre): cloud based NLP used for mental health research
KOMENTI (Birmingham): semantic query and information extraction framework
EdIE-Viz (Edinburgh): rule-based and neural network-based information extraction systems for brain radiology reports
HELIN (Health Entity Linking) pipeline: automated coding of patient-centric entities using neural natural language processing, and COMETA: a training set from Reddit data (Cambridge)
Use cases
Practical experience in benchmarking NLP tools for radiology (Edinburgh)
Automated medical coding and weakly supervised rare disease identification from clinical notes (Edinburgh)
Open-source tools and resources including code can be found on the HDR UK Innovation Gateway and HDR UK Text Analytics github repository. For more information about the tools and how you can use them for your research, contact Natalie Fitzpatrick.
Summary of Key Themes
There is a huge opportunity to use text analytics to improve research, hospital service planning and clinical decision-making. Challenges remain in navigating regulatory access to free text EHR data and deploying consistent, replicable methods.
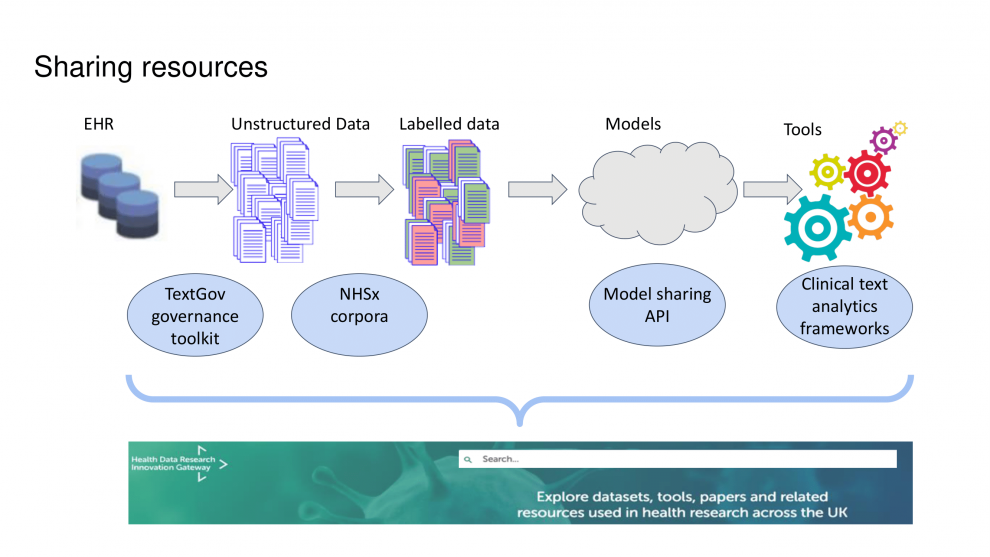
Data sharing and re-use of information
A key theme throughout the day was around how we can better share our tools and data including the creation of toolkits to enable people to share their resources across sites. Three levels of sharing as a community were discussed:
(1) Sharing tools, models and frameworks so people can re-use what has already been developed
(2) Sharing data and resources (e.g. training) for future NLP development
(3) Sharing common pathways and best practice and learning (e.g. Generation Scotland, EdIE-viz/EdI-R, CRIS, CogStack and developing corpora) is as important as model sharing
HDR UK is doing key work on bringing together all the tools and resources in clinical NLP via the Innovation Gateway. TexGov is developing data governance standards for using free text data in research and would be interested in hearing people’s own experiences and thoughts that could contribute to the development of these standards.
There was discussion around how we can extend sharing to include the commercial sector for mutual benefit. The question of how we get the tools that we generate in the NHS into the commercial sector was raised. Universities commonly ask commercial partners to partner up with UK university researchers and the university grants them honorary contracts.
Ethics approval
There was considerable interest in other people’s experiences of research ethics approval for projects utilising free text. Ethics committees responses seem heavily dependent on the experience of the particular committee in this area. For the THIN database-wide ethics approval including free text, the committee was interested in the entire process, with automated anonymisation treated as part of data minimisation rather than a definitive process to render the data de-identified. Other projects with generic ethics approval required researchers to provide a detailed explanation of why researchers wanted to develop NLP to their patient representatives.
It was felt that it would be extremely helpful to have access to a portfolio of successful ethics approved NLP studies that people would be happy to share with the community, including what are the key points that ethics committees found important to address, in order to share learning and understanding.
There was also interest in learning more about how researchers should approach ethics committees in relation to assessing NLP tools. For example, is there a pathway to apply for ethics to assess the NLP pipeline accuracy rather than for a specific patient outcome-centred research project?
Donating free text data for research
There was discussion around the usefulness of creating a bank of EHR from people who are happy to share their data. Goran Nenadic and team are putting together a group who are interested in clinical text donation which would fit in well with NHSX’s language corpora project. There could be a two stage anonymisation process involving a professional as well as a patient, and perhaps only text notes already shared with patients could be used.
The idea of getting patients involved in anonymising their own text was considered, although biases would need to be considered: EHRs contain data for patients who lack the capacity to anonymise text due to their illness or have a condition that prevents them from being able to do this. Other issues including how to develop NLP for sensitive topics that are redacted (such as safeguarding) and how to manage mention of third parties in text and how we can ensure that everyone is comfortable with what is going to be shared will need to be considered. NHS Trusts regularly have to screen records when patients ask for copies (to redact third party information) so this could follow a similar process. Despite this, it was felt that even a selected corpus of text could be useful for algorithm development. Ultimately it may be that a ‘mixed economy’ of text resources will be required. Donated text could result in major progress, but this will require broad representation.
Health data ownership, intellectual property and model disclosure
Ownership and IP rights is a complex area and a number of important questions that warrant further investigation were raised, including
• who 'owns' the health data and what consideration has been given to who owns the intellectual property (IP) of the developed NLP algorithms?
• as NLP models are trained on patient data, who owns the IP in the model?
• how do we ensure the benefits of using NLP are returned to the NHS?
• what contractual agreements are in place to protect against commercial companies profiting from NHS data?
• what would patients and public consider is excessive profiteering and what would they consider as fair profit?
The IP for research carried out in universities tends to belong to the university unless the research is being carried out as part of a start-up. However it is not just the IP of the algorithms and code, but also the models that are developed, that need to be considered.
IP should be contracted explicitly in service level agreements.
Disclosure of information within models needs careful thought (see below link on ''Generation and evaluation of artificial mental health records for Natural Language Processing'' in npj Digital Medicine volume) and robust approaches need to be considered (e.g. differential privacy and only training models on de-identified data). The government (?ONS) may have developed some guidance that may be a good starting point. MedCAT has two types of model. The model that does the named entity recognition and linking, the risk is extremely low as the model does not generate text. The meta-annotations model, does have language generation capability so theoretically is higher risk however the tokenizer (which is required to generate words) was not trained on patient sensitive data so the practical risk is very low. The model would also require significant refactoring to generate text, so would require an explicit “attack” rather than happening by accident. Google AI recently released a report on Privacy Considerations in Large Language Models which highlighted weaknesses in large language modelling.
Anonymisation
There was interest around what tools are best for de-identification. A sensible approach taken at King's College Hospital was to start really small, get tools installed and then start demonstrating some use cases and proof of concepts that really demonstrate value for patients that gives patients something tangible that is understandable. How data are anonymised is important. See Hercules Dalianis’s group’s work on de-identification strategies and their effect on downstream named entity recognition in clinical text.
Patient and Public Involvement
Involving patients and public in our work and understanding more about what patients and public want their data to be used for, and how, is important. Patient and public involvement in decisions about the use of their data is a key component of the work of TexGov (which is working towards the creation of data governance standards to enable free text data to be used safely for research for the benefit of patients and the public).
Next steps
• Explore, as a community, how we can extend sharing to include the commercial sector for mutual benefit
• Collate and make available examples of successful ethics applications for NLP studies that people can access, including what are the key points that ethics committees wanted addressed, in order to facilitate/expedite the process of applying for ethics approval
• Further exploration of which tools are best for de-identification
• Further discussion around the creation of a bank of free text EHR donated by patients for research, including how to address biases
• Further discussion amongst stakeholders including the commercial sector around health data ownership, intellectual property (including IP for model development) and model disclosure is warranted so the rules are clear
• Development of a plan for how the public can be involved in free text research as collaborators and research team members, and how to include public members on decision making panels (including what information they need about our research)
• Further understanding around what patients and public consider is excessive profiteering and what would they consider as fair profit
• More communication around how to access open-source tools and resources for the community
Links
- Read "Giving something back": A systematic review and ethical enquiry into public views on the use of patient data for research in the United Kingdom and the Republic of Ireland in Wellcome Open Res
- Read ''Should free-text data in electronic medical records be shared for research? A citizens’ jury study in the UK'' in BMJ
- Read about the TextGov project
- Go to HDR UK Innovation Gateway
- Read further on HDR UK Text Analytics github repository
- Read the paper about Disclosure of Information in npj Digital Medicine volume
- Check the Privacy Considerations in Large Language Models
- Read about The impact of de-identification on data quality in ACL Anthology
- Watch the Zoom recording of the workshop
- Email Natalie Fitzpatrick
 Close
Close

