Target Identification and Validation in the translation of small molecules involves identifying biological molecular structures and conducting validation experiments to show therapeutic effect.
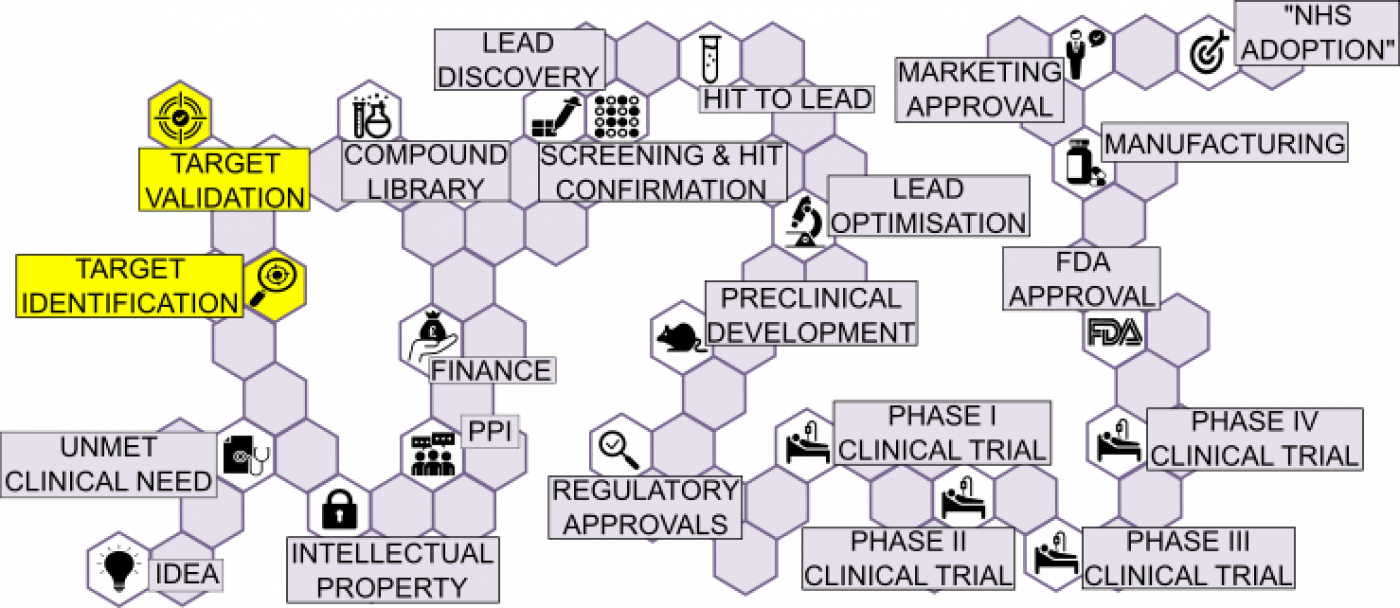
 TARGET IDENTIFICATION:
TARGET IDENTIFICATION:
Identifying biological molecular structures which are ‘Druggable’. Druggable biological targets interact with the small molecule to change or alter the function of the target.
Properties of an ideal target:
- The target has a pivotal role in the pathophysiology.
- The target expression is confined to a specific locations within the body.
- Enabled by a 3D model of the target for Druggable assessment.
- The target can be easily assayed in an high throughput screen.
- Regulation of the target by a small molecule has a favourable toxicity profile and potential side effects can be predicted using phenotypic data.
Approaches to target identification include:
- Phenotypic screen approach
- Genetic association studies.
 TARGET VALIDATION:
TARGET VALIDATION:
A prospective drug target has to undergo many validation experiments to show that it is directly involved in the pathway and that it will have a therapeutic effect.
How to validate a target:
- Druggability assessment – This involves exploring the sequence-related properties of the proteins and the 3D-sturucture using x-ray crystallography.
- Assayability assessment – This is to help with future compound screens. Biochemical and cellular assays need to be developed to enable the search for small molecule modulators of the protein.
- Genetic assessment - This involves looking at the genetic sequence and patient genetic material, which may predict efficacy of a potential therapy and predict potential adverse side effects. Targets may be expressed in different organs and even possess different functions dependent upon locations. It is important to know the expression profile of the target from human genetic data as it confirms that the target is implicated in the disease and this can be replicated in animal models.
ASSAY VALIDATION:
After identifying a target, a simple experiment to analyse enzyme activity or cell phenotype must be generated to screen against a small molecule library. This ‘Primary assay’ will be used to screen as many compounds as possible – potentially millions – so must meet a assay validation threshold before being used.
- Primary assay – This could be either a biochemical or cell based assay depending on the targeted disease. The assay would need to be HTS compatible – meeting the following criteria: potential for 1536-well plate format, demonstrable assay quality measures (assay window and robustness), known enzyme kinetics and reaction kinetics, demonstrated reagent and signal stability, DMSO tolerance, relatively inexpensive.
- Othorganol assay – A second assay that is performed after the primary assay, using an alternative substrate or technology platform. It is used to triage primary assay hits and remove false positives. 384-well format and can be more expensive as screen will be smaller.
- Target engagement assay – An assay to measure the extent to which the small molecules bind to the desired target. Depending on the available technology this strategy may be used as a Primary assay or on fewer compounds following triage of hit compounds.
UCL Support:
The TRO Drug Discovery Group (DDG) at UCL can help generate High Throughput Screening compatible assays.
With the aid of state-of-the-art screening capabilities and automated equipment, the DDG are able to handle high throughput screens of compounds in multiple disease settings quickly and efficiently. The Drug Discovery Group labs are capable of medium to high throughput biochemical and cellular screening in all major read modes.
 Close
Close

