MDC publication highlights – September 2020
17 October 2020
Clinical trial to evaluate Ursodeoxycholic acid (UDCA) as a disease-modifying treatment for Parkinson’s, How gene-environment interactions affect the risk for Parkinson’s & What can gene expression patterns teach us about the biological basis of dystonia?
Clinical trial to evaluate Ursodeoxycholic acid (UDCA) as a novel disease-modifying treatment for Parkinson’s
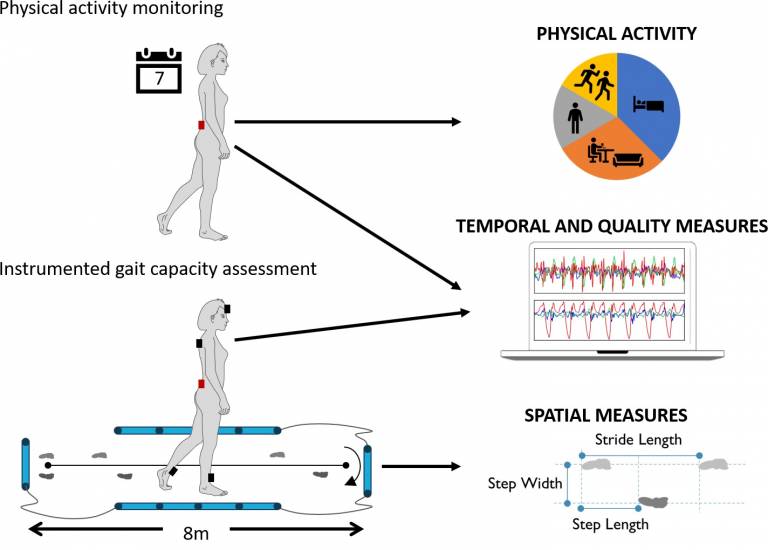
The development of treatments that could stop or slow Parkinson’s disease progression is a crucial step in reducing mortality and delaying the onset of severe complications associated with late-stage Parkinson’s. This article describes the ongoing clinical trial to tests the effects of a drug called Ursodeoxycholic acid (UDCA) on Parkinson’s disease progression. UDCA has been in clinical use for decades, primarily to treat liver disease, and is widely established as a safe and well tolerated drug. Earlier experiments in animal- and cell-based models indicated that UDCA could prevent the death of brain cells associated with Parkinson’s, likely doing so through its protective effect on the mitochondria; the cells’ energy-producing organelle. The research team, led by Prof. Oliver Bandmann from the University of Sheffield, in collaboration with Prof. Tom Foltynie from UCL MDC, are currently conducting a clinical trial to explore the effects of UDCA on people with Parkinson’s. The trial recruited 30 people who were diagnosed with Parkinson’s up to three years prior to starting the trial. Participants are treated with UDCA or an inactive placebo for one year, during which researchers will evaluate whether the drug has any adverse effects, and whether it has any beneficial effects in delaying the progression of Parkinson’s symptoms. The trial is using a novel brain imaging technique (Phosphorus MR spectroscopy) to assess the effects of UDCA on its molecular targets in the brain, as well as applying advanced wearable sensor-based technology for the objective quantification of motor symptoms. The trial is currently ongoing, with outcomes expected to be available by July 2021.
Payne et al. ‘Ursodeoxycholic acid as a novel disease-modifying treatment for Parkinson’s disease: protocol for a two-centre, randomised, double-blind, placebo-controlled trial, The 'UP' study’. Neurology, First Published: 5 August 2020, DOI: 10.1136/bmjopen-2020-038911.
How gene-environment interactions affect the risk for Parkinson’s?
The primary pathology leading to Parkinson’s disease (PD) is the death of dopamine-producing neurons, specifically in a part of the brain called the substantia nigra pars compacta. Evidence suggests that the loss of dopaminergic cells is a long and gradual process, likely spanning many years. However, clear clinical symptoms leading to a diagnosis of PD only start manifesting towards the end of this process, by which time as many as 90% of the dopamine-producing neurons will have been lost. Effective treatments that could slow or stop Parkinson’s disease progression that have the greatest chance of success are those aimed at preventing cell death, and thus must be administered early on, before the majority of neurons have been lost. It is therefore absolutely vital to develop ways of identifying people who are at greater risk for PD, even before the appearance of clear motor symptoms.
Over the past decade, large genetic studies have identified a number of genetic markers which indicate an increased risk for PD. Separately, epidemiological studies have identified a range of “environmental” (i.e., non-genetic) factors that could also influence the risk for PD, including life style factors, exposure to certain substances and others. However, the relative contribution of each individual genetic or non-genetic factor to the overall risk for PD is small, suggesting that interactions between them may explain some of the missing risk.
In this study, researchers from Wolfson Institute of Preventive Medicine in London and UCL MDC used data from the UK Biobank and most recent large genetic data in PD to investigate the association between environmental and genetic risk factors. The researchers sought to evaluate how the integration of these factors could improve the accuracy of predictive algorithms designed to identify at-risk individuals in the “prodromal” (pre-symptomatic) stage of PD. Outcomes of the study confirmed the association between PD and several well-established risk factors, such as having a family history of PD or dementia, not smoking, low alcohol consumption and others. Next, the researchers showed that integrating genetic data into a risk prediction algorithm based on environmental factors was able to improve the accuracy of the model, but the added value of the genetic data was very small. However, a direct investigation of gene–environment interactions uncovered a novel finding, whereby the effect of diabetes on PD risk appears to depend on background genetic risk. These findings could have implications for risk stratification of individuals for studies examining the ‘pre-diagnostic’ phase of PD and for our understanding of how genetic variation and other risk factors interact in the pathogenesis of PD.
Jacobs et al. ‘Parkinson’s disease determinants, prediction and gene–environment interactions in the UK Biobank’. Journal of Neurology, Neurosurgery and Psychiatry, First published: 15 September 2020, DOI: 10.1136/jnnp-2020-323646.
Using gene expression patterns to uncover the biological basis of dystonia
Dystonia is a neurological disorder characterized primarily by involuntary muscle contractions causing abnormal movements and postures. However, dystonia does not only affect motor function, but in a large majority of patients is also associated with psychiatric symptoms, such as depression, anxiety, psychosis and others. The biological mechanisms underlying many forms of dystonia, as well as its association with psychiatric symptoms, are largely unknown. A growing number of gene mutations have been identified in recent years that are linked to various forms of dystonia. However, very little is known about the biological function of these dystonia-related genes and how they contribute to the pathological process leading to dystonia. The current study, led by researchers from UCL Institute of Neurology and UCL MDC, explored the biological function of dystonia genes and their relationship to genes associated with various psychiatric disorders. Specifically, the researchers tried to identify which brain regions and cell types show the highest expression of dystonia genes, which cellular pathways might be affected by mutations in these genes and whether dystonia a genes and genes associated with psychiatric disorders have a shared biological basis. To answer these questions the researchers generated network models of dystonia gene expression, based on large databases of gene expression in specific brain tissues and cell types in mice and human brain. They identified three major brain regions that were enriched for expression of multiple dystonia-related genes, including dopamine-producing neurons in the substantia nigra, medium spiny neurons in the striatum and neurons in the frontal cortex. The researchers also found that many of these genes were related to synaptic function in neurons. Genes related to neuropsychiatric disorders were found to have some common expression patterns with dystonia genes, specifically in the putamen and white matter regions. These results indicate that dystonia-associated genes likely contribute to pathogenesis of dystonia through dysregulation of synaptic signalling in specific brain regions (striatum, substantia nigra and frontal cortex). Based on shared patterns of genes expression, it is likely that the same biological dysfunction underlies both dystonia and the associated psychiatric symptoms.
Mencacci et al. ‘Dystonia genes functionally converge in specific neurons and share neurobiology with psychiatric disorders’. Brain, First published: 21 August 2020, DOI: 10.1093/brain/awaa217.
 Close
Close

