TCR repertoires
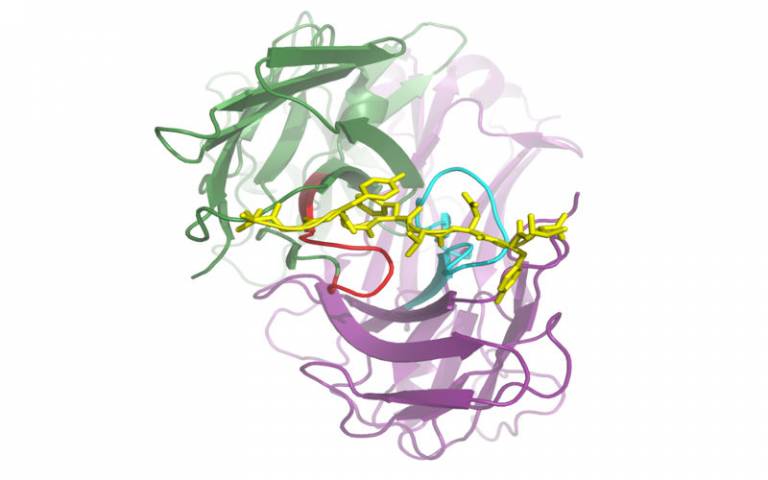
An individual's immune system contains a large number of T-cells expressing a large number of TCRs which can respond to different pMHCs. The repertoire of different TCRs that an individual's immune system contains thus in some way relates to the number of different antigens that the system can respond to. As T-cell clones divide and contract in response to the presence or removal of their antigen, there is further information in a repertoire relating to past and ongoing immune responses in that individual.
We therefore believe that by sampling and measuring the repertoire of TCRs in a blood or tissue sample one should be gaining access to a tremendously powerful biomonitor, a readout of the past, present and potentially future status of that immune system. However due to the recombinatorial and non-templated base editing processes that occur during TCR production in the thymus there is an enormous number of different TCRs, with theoretical estimates of total potential diversity far exceeding the number of cells in a human body.
Therefore we in the Innate2Adaptive lab have developed a number of pipelines and protocols for the unbiased, high-throughput amplification, sequencing and analysis of TCR repertoires. These approaches include using unique molecular barcoding error-correction procedures to produce robustly quantitative repertoire data, which is analysed using the Decombinator suite of analysis scripts that we have developed for the rapid identification and description of rearranged TCR sequences.
We are interested in using our TCR sequencing pipeline to explore T-cell biology in many different settings, including investigating the perturbance of repertoire features in response to infectious challenge, assessing the impact of primary immunodeficiencies, and exploring the clonal distribution between different T-cell subsets and tissues.
 Close
Close

