Plants and Crops
The UCL Research Department of Genetics, Evolution and Environment undertakes a broad range of research into plants, and particularly crops.
The evolution of plants via natural selection and the improvement of crops by artificial selection represent two sides of the same coin, and our research programmes in GEE explore both aspects. We use a combination of empirical studies of plant evolution (Jon Bridle), ancient crop plant DNA (Hernán Burbano and Richard Mott) and experimental intercrosses (Richard Mott) to dissect the genetics of adaptation and domestication and improvement.
This work is particularly relevant given the important challenges induced by climate change on natural ecosystems and on farming.
Plant responses to environmental change: investigating the evolution of plant behaviour at ecological margins

Our research compares evolutionary responses to ecological variation along an elevational gradient on Mount Etna, Sicily.
How plants respond to changing environments is a fundamental question both in understanding the resilience of ecological communities, and in the maintenance of crop yields in increasingly stressed environments. Our research group, headed by Jon Bridle, uses a combination of genomic, ecological, and quantitative genetic analyses to test limits to the adaptive plasticity of two Senecio ragwort species, which show recent rapid ecological divergence to an elevational gradient.
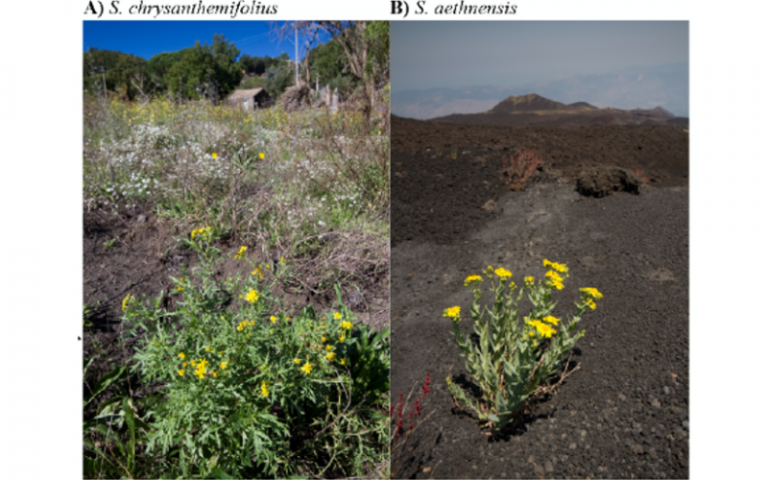
Two closely related species of Senecio ragworts are each restricted to low (A) and high (B) elevations, and show distinct leaf morphology, physiology, and life history related to their distribution.

Prof Antonia Cristaudo (University of Catania) taking cuttings from wild genotypes for propagation in the greenhouse.

With collaborators at the Universities of Oxford, Catania, Monash and Napoli, we are conducting large scale transplant experiments to test the limits of these species' behavioural responses within and beyond their current ecological range.
Analyses of transcriptomes of field transplanting cuttings also allow exploration of the genomic architecture of plant behaviour, both between these species during their recent adaptive divergence, as well as within genotypes across their native elevational range and beyond. Work is ongoing to relate such shifts in gene networks to changes in trait correlations and trade-offs across these environments, in order to understand the limits of natural and artificial selection in effecting phenotypic and
behavioural evolution.

Transplanting cuttings at our 2000 m site on Mount Etna, May 2019
Burbano Lab
My group uses a retrospective approach to address a broad range of questions in the evolution of plants and their pathogens. This approach gains its power from obtaining direct observations from the past through sequencing historical and ancient samples, and combining them with present-day genomic information. My group has pioneered the use of herbarium specimens and plant archaeological remains for evolutionary genomics studies. Currently, our research focuses on three main topics:
The dynamics of present and past plant-pathogen epidemics: The study of the dynamics of past epidemics and the co-evolution of plant-pathogen interactions is of major relevance, since the emergence and re-emergence of plant diseases pose a major threat to food security and indirectly to human health. Additionally, the strength of natural and artificial selection experienced by the plant-pathogen system permits observing changes in allele frequencies in short periods of time.
My group has centered on the study of 19th and 20th century outbreaks caused by Phytophthora infestans, the causal agent of potato late blight (Yoshida et al. 2013, eLife; Yoshida et al. 2014, PLoS Pathogens) and by the rice blast fungus Magnaporthe oryzae (Latorre et al. 2020, BMC Biology).
A potato specimen from the Kew Garden herbarium, collected in 1847, during the height of the Irish famine. © Marco Thines/Senckenberg Gesellschaft für Naturforschung
The identification and timing of key events in crop domestication and the spread of agriculture: Using an approach that combines ancient genomics with quantitative and population genetics, my group started characterizing the spread of maize in North America and its adaptation to a temperate climate. Using techniques borrowed from breeding (genomic selection) we predicted for the first time polygenic quantitative traits on archaeological remains (Swarts et al. 2017, Science).
The post-domestication dispersal of crops domesticated in the Americas presents a unique opportunity to study the population history and adaptation of plants. Our group reconstructed in great detail the population history of European potatoes combining present-day and historical genomes. Our work showed how allele frequencies can be used under an admixture graph framework to study the relations between temporally and spatially diverse populations (Gutaker et al, 2019, Nature Ecology and Evolution).
A potato (Solanum tuberosum) herbarium specimen collected in Chile by Charles Darwin on the voyage of the Beagle. © Cambridge University Herbarium.
The colonization of new ecological niches by invasive/introduced species: Our group has explored the colonization of North America by Arabidopsis thaliana using a dense time-series spanning the 19th and 20th centuries to examine the evolutionary forces that have shaped genetic variation in a recently introduced species. We estimated the long-term nuclear mutation rate and showed that although genetic drift predominates, purifying selection also shapes diversity in this colonizing population (Exposito-Alonso et al., 2018. PLoS Genetics).
Richard Mott Group
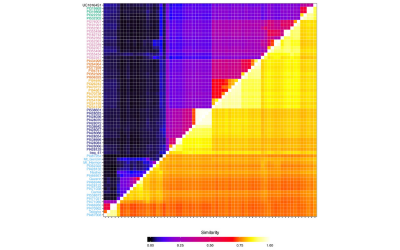
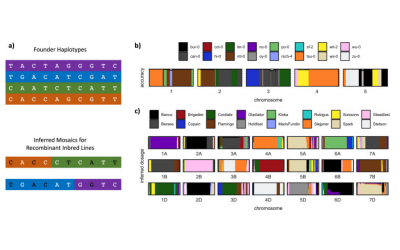
1. We carry out research in multiparental populations. These are panels of recombinant inbred lines descended from multiple inbred founders (MAGIC: Multiparent Advanced Generation Inter Cross). We work on the Arabidopsis, rice and wheat MAGIC populations. We develop statistical methods for mapping quantitative trait loci and for imputing genomes from low-coverage sequencing, and we have sequenced each of these populations. Most of this work has been funded by the BBSRC and GCRF.
(a) The Arabidopsis MAGIC was developed in collaboration with Paula Kover (University of Bath). We sequenced the 19 founders of the population in one of the earliest pan genome studies. We have used low-coverage sequence of the population to show how to map structural variations.

Seedlings of accessions of Arabidopsis thaliana
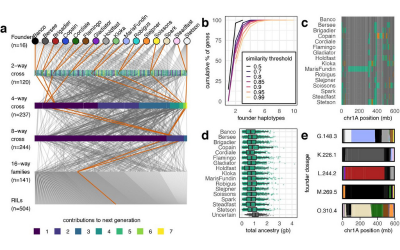
(b) The DIVERSE Wheat MAGIC was developed in collaboration with James Cockram and colleagues at the National Institute for Agricultural Botany (NIAB), Cambridge. The population was designed to sample the major varieties grown in the UK over the past 70 years. We have sequenced the 16 founders by capture and sequenced over 500 genomes at low coverage and mapped over 130 quantitative trait loci in the population.
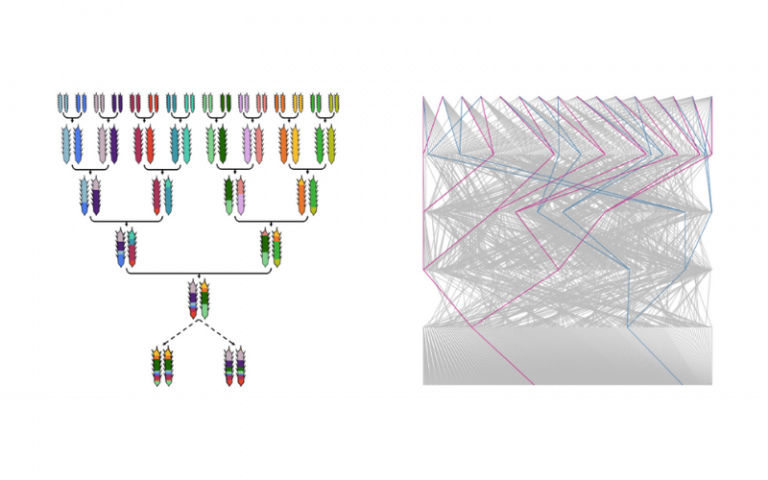
Crossing scheme for the Diverse Wheat MAGIC

The Diverse Wheat MAGIC

The Diverse Wheat MAGIC
(c) The rice HEAT MAGIC was developed at the International Rice Research Institute, Philippines, to identify loci associated with heat tolerance. We have sequenced over 800 lines at low coverage, which have been used to identify loci associated with chalkiness.

Rice MAGIC grown at IRRI, Philippines

Rice fields at IRRI, Philippines
2. We are collaborating with Rajeev Varshney and colleagues at the International Crop Research institute for the Semi-Arid Tropics (ICRISAT, India) on chickpea population sequencing. We have sequenced 2000 landraces of chickpea at low coverage.

ICRISAT, India

Automated chickpea phenotyping, ICRISAT, India
(3) We are interested in ancient crop genomes. In collaboration with Mark Thomas, Dorian Fuller and colleagues in UCL Dept of Archaeology and the UCL Museum of Egyptian Archeology we sequenced a 3,000 year old specimen of emmer wheat from the Ramessid period and show that most of the genetic signatures of domestication were fixed by that date. It is genetically similar to modern emmer wheats that are grown in India, Oman, and Turkey.
Compared to wild relatives, domesticated wheats have larger grains that germinate readily; the seeds are also strongly attached to the plant (scar circled in picture), where they can be harvested more easily.

Ancient Egyptian Emmer Wheat at the UCL Petrie Museum of Egyptian Archaeology

Ancient Egyptian Emmer Wheat at the UCL Petrie Museum of Egyptian Archaeology
 Close
Close