PRMTs and the regulation of NSC proliferation and differentiation
Protein arginine methylation is a common post-translational modification. However, compared to other post-translational modifications, e.g. phophorylation, very little is known about how the enzymes responsible, the PRMTs, are regulated and what the biological outcomes of such modifications are. Recently, arginine methylation has
attracted more attention as it became clear that histones are arginine-methylated and that this modification has important consequences for gene expression. There are two major classes of PRMTs, type I and type II. Both types transfer the methyl groups from a donor molecule, S-adenosylmethionine (SAM), to the terminal guanidino nitrogens of arginine residues generating 1) mono-methyl 2) symmetric dimethyl and 3) asymmetric dimethyl arginine. Type I enzymes mediate asymmetric dimethylation, type II enzymes catalyze symmetric dimethylation. Generally, asymmetric dimethylation of histones leads to transcriptional activation while symmetric dimethylation leads to transcriptional repression. Genetic ablation of specific PRMTs in mice has highlighted their importance in early development and differentiation of multiple tissues. There is evidence that asymmetric dimethylation mediated by PRMT1, a type I enzyme, increases at the onset of differentiation, e.g. during erythroid differentiation and neuronal differentiation in nerve growth factor (NGF) treated PC12 cells. The significance of symmetric dimethylation mediated by the class II enzyme, PRMT5 is poorly understood. In the context of chromatin, PRMT5 methylates predominantly histones H4R3 and H2AR3 but can also methylate histone H3R8. PRMT5 null mice exhibit an early embryonic lethal phenotype highlighting its importance in early zygotic development. Recently, PRMT5 emerged as a vital regulator of cellular "stemness". For example, PRMT5 is essential for maintaining pluripotency of embryonic stem (ES) cells, regulates reprogramming of primordial germ cells (PGCs) during development and silences developmentally regulated gene expression in erythroid progenitors.
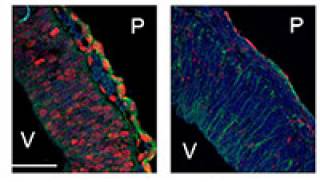
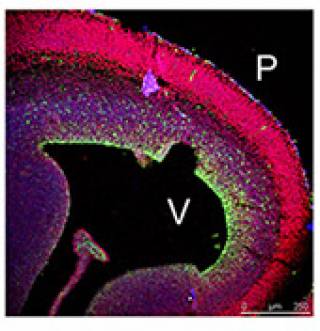
I recently showed that the specific histone modification mediated by PRMT5, H4R3me2s, is prevalent in the embryonic day 10.5 (E10.5) cerebral cortex, which is undergoing mainly symmetric "expansion" divisions, whereas PRMT1-mediated asymmetric dimethylation of the same residue, H4R3me2a, appears in post-mitotic neurons. NGF-induced neuronal differentiation of PC12 cells is dependent on PRMT1-mediated asymmetrical methylation. Moreover, the identification of TIS21, a stimulator of PRMT1 activity, as a marker of neuroepithelial cells that will generate a post-mitotic neuron or neuronal progenitor at their next division provides another link between neuronal differentiation and the activation of PRMT1 and its product H4R3me2a. I am interested in identifying the extracellular signals which regulate the activity of both PRMT5 and PRMT1 during neural development and identifying the substrates of these enzymes during neurogenesis and gliogenesis.
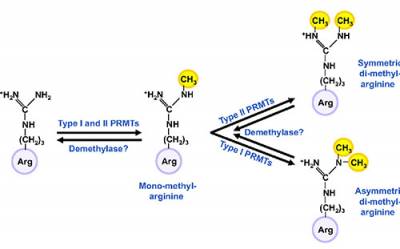
Fig. 1. Tudor domains bind methylated arginines and lysines. (B) PRMTs mediate either symmetric or asymmetric di-methylation of arginine.
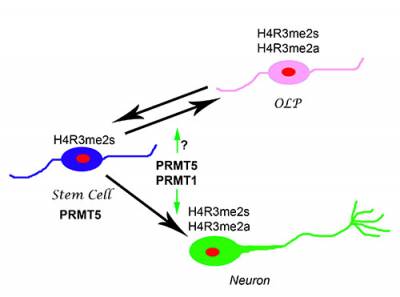
Working Hypothesis. PRMT5-mediated H4R3me2sym specified the histone code for the "stem-like" cellular state of the NSCs. Deposition of PRMT1-mediated H4R3me2asym modifications specify the commitment to neural differentiation
 Close
Close