PRDM4 and the PRDM Family of transcription factors
PRDM4 belongs to a family of related "PRDM" proteins which contain two distinct
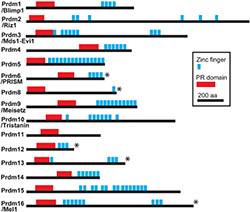
fig. 1 From Hohenauer T and Moore A, 2012, Development
structural domains: 1) the zinc finger (ZF) domain and 2) the positive regulatory/suppressor of variegation, enhancer of zeste, trithorax (PR/SET) domain characteristic of histone lysine methyltranferases (HMTases). PRDM proteins are transcriptional regulators of differentiation in multiple tissues. They recognize and bind to specific DNA sequences through their ZF domains. They function either as epigenetic chromatin modifiers in their own right or by recruitment of third-party chromatin modifiers to control transcription of lineage-specific genes. For example, PRDM1/Blimp 1, is a critical developmental regulator in many tissues, recruiting different chromatin modifiers in a context-dependent manner, instructed by cell-extrinsic signals. Moreover, deregulation of several PRDM proteins is associated with cancer, highlighting their role as tumour suppressors.
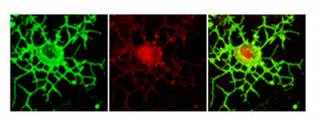
I originally identified PRDM4 as a protein that interacts with the low affinity neurotrophin receptor, p75NTR, implicating PRDM4 in neurotrophin-mediated control of neural development (Chittka and Chao, 1999). I showed that PRDM4 is a
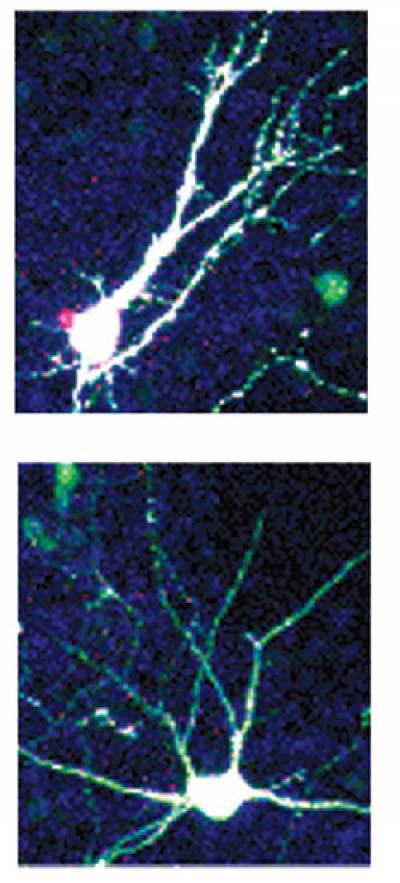
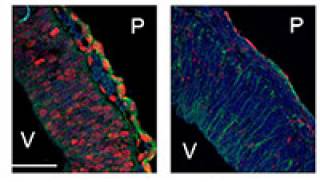
Knockdown of PRDM4 in NSCs leads to precocious neurogenesis. Transfected NSCs immunolabelled for EGFP and early neuronal marker, TuJ1 - merged staining.
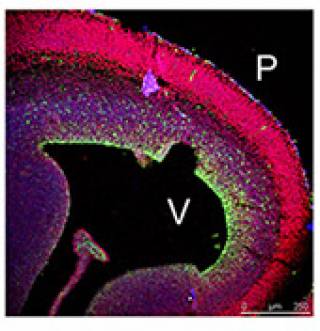
transcriptional repressor that recruits histone deacetylases (HDACs) 1 and 2, suggesting that it exerts a chromatin-mediated transcriptional activity (Chittka et al, 2004). PRDM4 is widely expressed in the developing CNS. My recent work addressed the role of PRDM4 in the regulation of cortical NSC development. I found that NSCs isolated from embryonic mouse cerebral cortex express PRDM4 and that it is down-regulated at the onset of neurogenesis. Knock-down of PRDM4 expression leads to precocious neurogenesis. Expression of PRDM4 persists in proliferating oligodendrocyte precursors (Chittka et al, 2012).
Moreover, I showed that PRDM4 exerts part of its biological activity by recruiting a chromatin modifier, the protein arginine methyltransferase, PRMT5 to mediate histone H4 arginine 3 symmetric dimethylation (H4R3me2s) and in this way to regulate gene transcription. My recent findings indicate that PC12 cells treated with the Epidermal Growth Factor (EGF) sustain high levels of PRDM4:PRMT5-mediated H4R3 symmetric di-methylation. Interestingly, in PC12 cells treated with the Nerve Growth Factor (NGF), PRDM4:PRMT5-mediated H4R3me2s rapidly decreases (Chittka, 2013). These observations and the fact that PRDM4, PRMT5 and H4R3m2s, are present in the undifferentiated neuroepithelium and dissociated NSCs during symmetric expansion divisions, support the general idea that PRDM4:PRMT5-mediated symmetric histone arginine methylation constitutes part of the developmental programme that specifies the "undifferentiated" cellular state.
 Close
Close