Overview
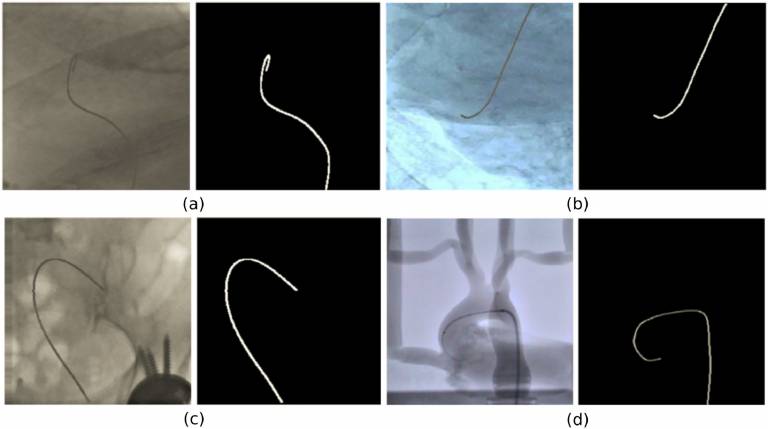
This dataset contains fluoroscopy images extracted from four videos of canulation experiments with an aorta phantom and six videos of in-vivo catheterisation procedures: four Transcatheter Aortic Valve Implantations (TAVI) and two diagnostic catheterisation procedures. Please refer to the README.docx
The Phantom.hdf5 file contains the 2000 (Dataset-2 in the paper) images extracted from the four fluoroscopy videos from catheterization experiments carried out on a silicon aorta phantom in an angiography suite.
The T1T2.hdf5 and T3-T6.hdf5 files contain images extracted from the six fluoroscopy videos during in-vivo endovascular operations (Dataset-3 in the paper). Specifically, 836 frames were extracted from TAVI (data groups T1, T2, T3 andT4) and 371 from diagnostic catheterization (data groups T5 andT6). Each data group contains the following number of images: T1 – 286, T2 – 150, T3 – 200, T4 – 200, T5 – 143, T6 – 228.
Binary segmentation masks of the interventional catheter are provided as ground truth. A semiautomated tracking method with manual initialisation (http://ieeexplore.ieee.org/document/7381624/) was employed to obtain the catheter annotations as the 2D coordinates of the catheter restricted to a manually selected region of interest (ROI). The method employs a b-spline tube model as a prior for the catheter shape to restrict the search space and deal with potential missing measurements. This is combined with a probabilistic framework that estimates the pixel-wise posteriors between the foreground (catheter) and background delimited by the b-spline tube contour. The output of the algorithm was manually checked and corrected to provide the final catheter segmentation.
The annotations are provided in the files: “Phantom_label.hdf5”, “T1T2_label.hdf5” and “T3-T6_label.hdf5”. All annotations consist of full-scale (256x256 px) binary masks where background pixels have a “0” value, while a value equal to “1” denotes the catheter pixels.
Example python code (MAIN.py) is provided to access the data and the labels and visualize them.
Downloading the dataset
If you wish you download this dataset, please visit HERE.
Citing the dataset
Please cite the following publication whenever research making use of this dataset is reported in any academic publication or research report:
Marta Gherardini, Evangelos Mazomenos, Arianna Menciassi, Danail Stoyanov, “Catheter segmentation in X-ray fluoroscopy using synthetic data and transfer learning with light U-nets”, Computer Methods and Programs in Biomedicine, Volume 192, Aug 2020, 105420, doi:10.1016/j.cmpb.2020.105420.
Contact
For comments, suggestions or feedback, or if you experience any problems with this website or the dataset, please contact Evangelos Mazomenos.
To find out more about our research team, visit the Surgical Robot Vision and Wellcome/EPSRC Centre for Interventional and Surgical Science websites.
 Close
Close

