Research
The Centre for Computational Science is committed to advancing science through computers. We do this in all ways we can, in the fields of physics, material sciences and chemistry to life, medical and clinical sciences.
At the CCS:
- We conduct research in several domains. These include lattice-Boltzmann fluid dynamics simulations (e.g., of blood flow, turbulent fluids, and liquid crystals), analysing drug effectiveness in patients suffering from HIV and cancer, modeling interactions between different materials (e.g. between clay sheets and oil drilling fluids), modeling DNA translocation and modeling vehicular traffic.
- We have developed a range of simulation codes to aid in our research and software tools to make production infrastructures easier to access and use. Hereby we aim to develop codes which run efficiently on the largest petascale architectures today as well as the exascale architectures which are expected to emerge in the near future.
- We build complex problem solving environments, including ones for clinical decision support.
Our different computational techniques span time and length-scales from the macro-, through the meso- and to the micro- and nanoscopic scale. We are committed to studying new approaches and techniques that bridge these scales.
We explore these domains through high performance, data intensive, super- and distributed (grid/cloud) computing, using resources within Europe as well as the United States.

Complex Fluids: The project features the world's largest lattice Boltzmann simulation of its kind, to study defect dynamics in liquid crystalline surfactant systems.
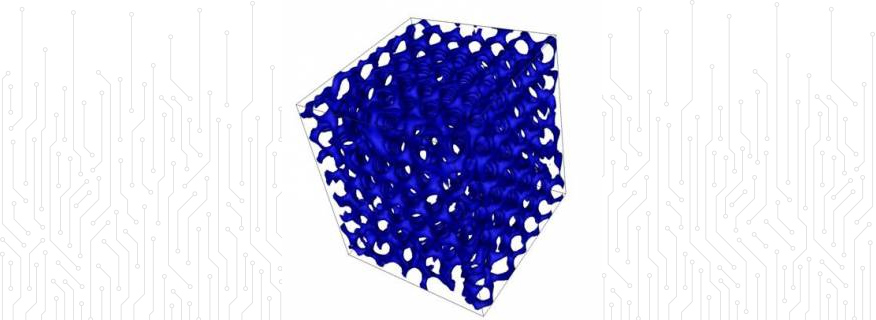
Complex Fluids: Lattice Boltzmann (LB) schemes have emerged in the past years as a valid tool for simulating the dynamics of complex fluids.
Complex Fluids: The lattice Boltzmann for Complex Fluids method is a mesoscale method for simulating complex fluids.
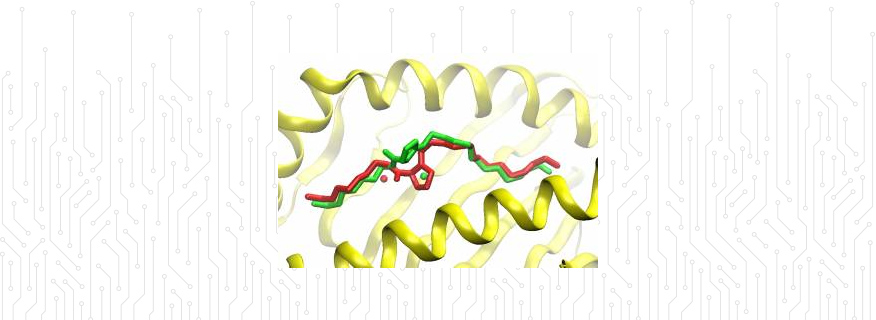
Biomolecular modelling: Understanding the interactions of epitopes bound to MHC proteins by T-cell receptors is an important step in the adaptive immune response and vaccines.
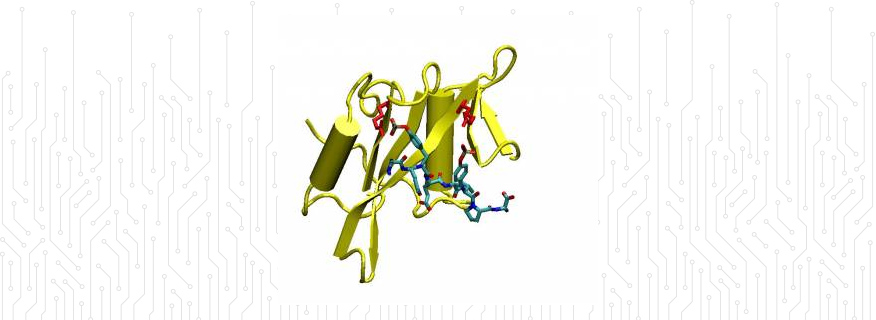
Biomolecular modelling: We are studying the binding of different peptide sequences to an SH2 domain using thermodynamic integration.
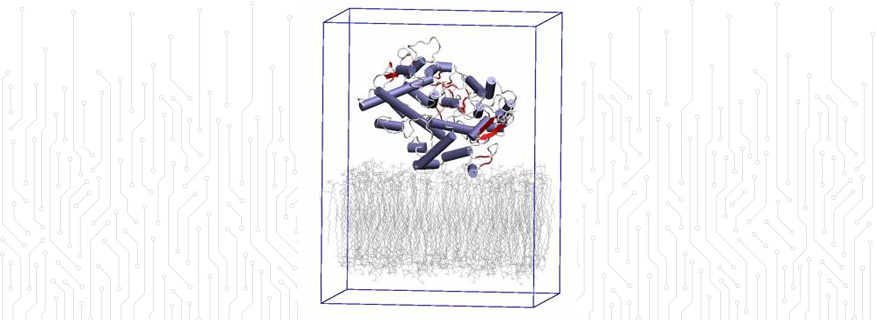
Biomolecular modelling: using large-scale classical molecular dynamics to study the dynamics between COX-1 and COX-2 for future NSAIDs development

Biomolecular modelling: HIV-1 Protease is an aspartic protease and is one of the key enzymes in HIV, essential for the maturation of new infectious virions
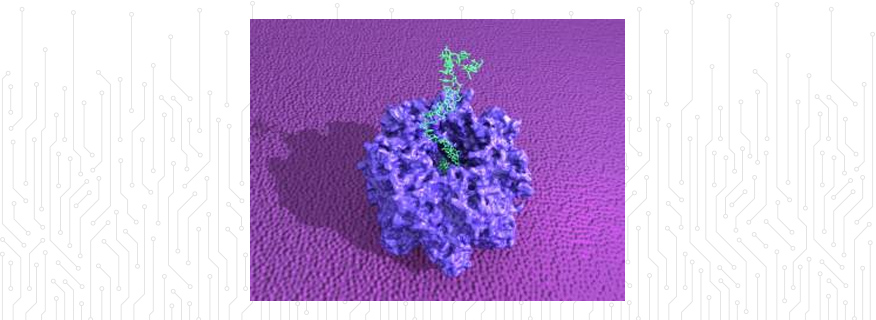
Biomolecular modelling: The translocation of polynucleotides through transmembrane protein pores is a fundamental biological process with important technological and medical relevance.
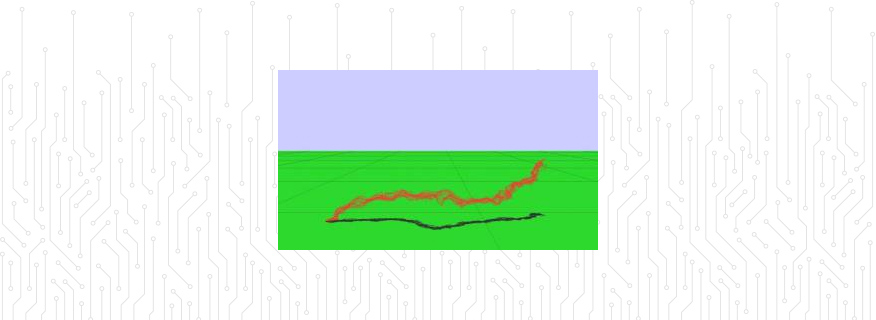
New methods for molecular simulations: We have developed and tested a new hybrid algorithm comprising molecular dynamics (MD) and computational fluid dynamics (CFD).
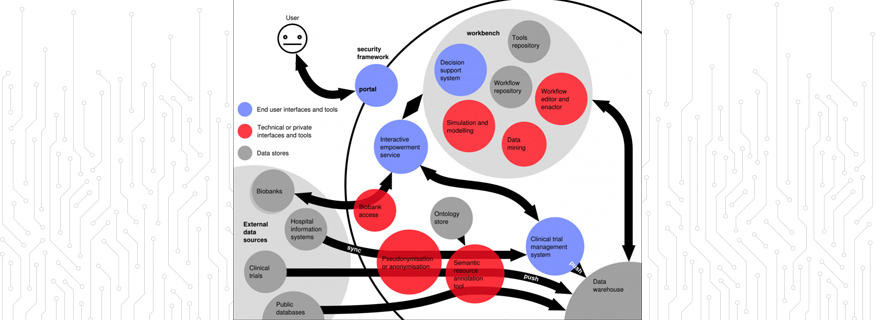
Data Handling: Health informatics relies, in part, upon computational simulation, modelling and data mining methods.
 Close
Close











