Editor – Ruth Lovering
Goodbye to Shirin Saverimuttu
Shirin has taken up a new role as a Trainee Polygenic Score Catalog Curator at EMBL-EBI after over a year as a member of the UCL Functional Gene Annotation Group. In this new role Shirin will be building on her curation experience and MSc in human genetics by extracting and curating information on polygenic risk scores from published articles. Shirin has been an asset to the team, not only providing excellent annotations, but also using her experience as a MSc student to improve our bioinformatics module and to train and guide last year’s project students in Gene Ontology annotation. I will miss her enthusiastic help and commitment and wish her the very best of luck in following a career as a biocurator.
Annotation progress
In total, all ARUK-UCL projects have resulted in 14,267 annotations for 2,658 distinct gene products, of which over 10,700 annotations are associated with 1,867 human gene products (data from QuickGO, 1 December 2020). Our current annotation focus is on Alzheimer’s disease (AD) susceptibility loci. Shirin and Penelope Garmini (EMBL-EBI curated funded by ARUK) have curated 13 of these through the review of 55 articles, providing over 200 annotations. For example, the role of complement C3b/C4b receptor 1 in cytolysis, immune complex clearance by erythrocytes and regulation of T-cell proliferation. Over the past year Ruth has been working with Pascale Gaudet, Swiss Institute of Bioinformatics, and Colin Logie, Radboud University, to improve the curation of DNA-binding transcription factors (dbTFs). In October the review of the literature supporting the assignment of dbTF activity to over 2000 proteins was completed with the result that 1,500 human proteins are now associated with the GO term ‘DNA-binding transcription factor activity’ or one of its descendant terms. The UCL team provided annotations for almost 200 of these proteins.
MSc annotation projects
This year a BHF PhD student, Kan Yan Chloe Li, is undertaking an annotation project, describing the key proteins involved in sinoatrial and atrioventricular node development. While the two iBSc students that have joined the team, Angeline Pesala and Praoparn Asanitthong, will be curating microRNAs, with a focus on those microRNAs that target proteins with a role in foam cell formation and atrial fibrillation (respectively).
Virtual meetings attended
November was a busy month, during which Ruth participated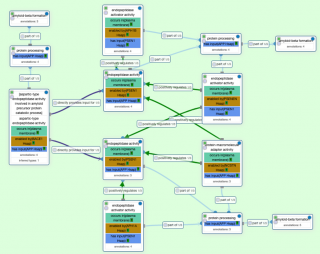
Recent Publications
- Lovering RC, Gaudet P, Acencio ML, Ignatchenko A, Jolma A, Fornes O, Kuiper M, Kulakovskiy IV, Laegreid A, Martin MJ, Logie C. A GO catalogue of human DNA-binding transcription factors. bioRxiv 2020 2020.10.28.359232.
- RNAcentral Consortium. RNAcentral 2021: secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2020 Oct 27:gkaa921. Epub ahead of print. PMID: 33106848.
- Wood V, Carbon S, Harris MA, Lock A, Engel SR, Hill DP, Van Auken K, Attrill H, Feuermann M, Gaudet P, Lovering RC, Poux S, Rutherford KM, Mungall CJ. Term Matrix: a novel Gene Ontology annotation quality control system based on ontology term co-annotation patterns. Open Biol. 2020 Sep;10(9):200149. PMID: 32875947.
 Close
Close

