Editor - Ruth Lovering
Gene annotation
BHF-UCL submitted a total of 720 annotations to 140 proteins to support to the second 'Critical Assessment of protein Function Annotation experiment'. The BHF team supported this experiment by manually curating proteins included in CAFA2, which are also on our cardiovascular priority gene list. The manual annotations we submitted can now be compared to the predicted annotations generated by CAFA2.
Based on the EBI statistics, 5th July 2014, this project has associated almost 31,000 GO terms to 4,300 proteins, 21,352 of which are to 2,448 human proteins. Note that the numbers of annotations created by this project are currently being displayed in two rows in the current composition table at the EBI: 'Manual GO annotation by BHF-UCL' and 'Manual GO annotation by BHF-UCL for Automated Function Prediction SIG'. This is because we are contributing annotations to support to the second 'Critical Assessment of protein Function Annotation experiment' (these are the CAFA2 annotations).
In addition, we have submitted 846 protein-protein interactions (PPIs) to IntAct, from the curation of 124 papers, which are now exported to public PPI databases. Since May we have completed the re-annotation of papers describing lipid metabolism, thanks to the hard work of Nancy and Mila. Currently in IntAct there are 1578 PPIs associated with the proteins encoded by the genes adjacent to the 82 genetic loci associated with total cholesterol levels (Teslovich et al. 2010), 6% of these were submitted by the BHF curators.
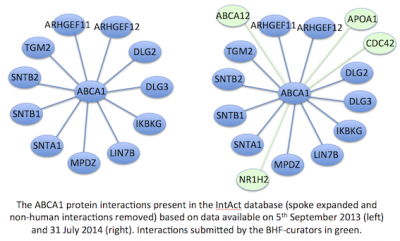
In some case these additional PPIs have substantially improved the networks available for individual proteins, for example the number of PPIs associated with ABCA1 has increased by 30%, from 11 PPIs to 15 (see figure above). Mila has now started capturing PPIs from papers describing heart development. To view the PPIs associated with a specific protein visit the IntAct browser, and type your protein of interest in the search field, a gene symbol will retrieve PPIs in a variety of species, whereas a UniProtKB ID will retrieve only the PPIs relevant to that protein. Suggestions for additional missing annotations are welcome, and should be sent to goannotation@ucl.ac.uk.
The EBI Proteomic Services have released a news item describing how bioinformatics tools can help the study of cardiac disease. This takes a closer look at KCNH2, its expression and the networks and pathways it is associated with, and includes annotations submitted by the BHF team.
Upcoming meetings
In September Ruth will be attending the British Atherosclerosis Society meeting, which has a focus on systems biology this year, and then the GO Consortium meeting in October.
Publications
A method for increasing expressivity of Gene Ontology annotations using a compositional approach. Huntley RP, Harris MA, Alam-Faruque Y, Blake JA, Carbon S, Dietze H, Dimmer EC, Foulger RE, Hill DP, Khodiyar VK, Lock A, Lomax J, Lovering RC, Mutowo-Meullenet P, Sawford T, Van Auken K, Wood V, Mungall CJ. BMC Bioinformatics. 2014 May 21;15:155. PMID:24885854
Contact goannotation@ucl.ac.uk to subscribe to this quarterly Newsletter
 Close
Close

