Welcome to the Jasmin Fisher Lab.
Group Leader: Jasmin Fisher, PhD, FRSB
Professor of Computational Biology
Research focus and mission
Our lab develops state-of-the-art computational models and analysis techniques to study cancer evolution and mechanisms of drug resistance to identify better personalised treatments for cancer patients.
Our research is focused on understanding how cancers evolve through the identification of molecular mechanisms that underpin cell-fate decision programs during both normal development and disease. We have developed an innovative approach called Executable Biology, which is a toolset to simulate and analyse biological mechanisms as if they were computer programs. This approach enables us to use powerful methods developed in computer science to prove properties of these programs and simulations to gain better understanding of the dynamic complexity of evolving biological processes such as cancer. This approach has been shown to be highly effective in developing deeper insights into the molecular mechanisms of cell fate decisions and in the discovery of novel combination therapies for cancer.
Our goal is to determine the mechanistic programs by which oncogenic signalling pathways regulate the onset, progression, maintenance and (when blocked) regression of cancers. We do this by computational modelling of oncogenic signalling networks and how they are linked to cell fate decisions (such as proliferation and cell death). In this way, we can understand how cellular decisions are made and how aberrations in those decisions drive the pathology of cancer.
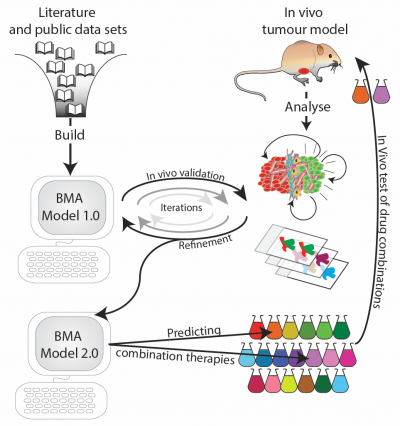
Iterative cycle between experimental lab work and computational modelling.
PNAS 116(44): 22399-22408, 2019
Our Lab
After more than a decade in the Department of Biochemistry at the University of Cambridge, our lab has recently moved to the UCL Cancer Institute to strengthen our collaborations with the outstanding talent of medical and clinical oncologists and cancer biologists within the institute.
Our multidisciplinary lab hosts students, postdocs and visiting researchers from a diverse set of backgrounds (e.g., biology, physics, computer science, software engineering, medicine) all working together as a team. We collaborate closely with experimental cancer biologists and clinicians to build and analyse our computational models and validate their predictions experimentally. When appropriate, we aim to translate our findings into clinical trials to improve cancer patients’ outcome.
Selected publications
Howell R., Davies J., Clarke M., Appios A., Mesquita I., Jayal Y., Ringham-Terry B., Boned Del Rio I., Fisher J.* and Bennett C.
Localized immune surveillance of primary melanoma in the skin deciphered through executable modellingScience Advances, 9:15, 2023- Howell R., Clarke A.M., Reuschl A.K., Chen T., Abbott-Imboden S., Singer M., Lowe D.M., Bennett C.L., Chain B., Jolly C. and Fisher J.
Executable Network of SARS-CoV-2-Host Interaction Predicts Drug Combination Treatments.
npj Digital Medicine, 5:18, 2022
- Clarke M.A. and Fisher J.
Executable Cancer Models: Successes and Challenges
Nature Reviews Cancer 20: 343-353, 2020
- Kreuzaler P., Clarke M.A., Brown E.J., Wilson C.H., Kortlever R.M., Piterman N., Littlewood T., Evan G.I. and Fisher J.
Heterogeneity of Myc expression in breast cancer exposes pharmacological vulnerabilities revealed through executable mechanistic modelling
Proc Natl Acad Sci U S A 116(44): 22399-22408, 2019
- Yu M.K, Ma J., Fisher J., Kreisberg J.F., Raphael B.J. and Ideker T.
Visible Machine Learning for Biomedicine
Cell 173: 1562-1565, 2018
- Silverbush D., Grosskurth S., Wang D., Powell F., Gottgens B., Dry J.R. and Fisher J.
Cell-Specific Computational Modeling of the PIM pathway in Acute Myeloid Leukemia
Cancer Research 77(4), 2017
- Chuang R., Benque D., Cook B., Hall B.A., Ishtiaq S., Piterman N., Taylor A., Vardi M., Koschmieder S., Gottgens B. and Fisher J.
Drug Target Optimization in Chronic Myeloid Leukemia Using an Innovative Computational Platform
Nature Scientific Reports 5:8190, 2015
- Moignard V., Woodhouse S., Haghverdi L., Lilly J, Tanaka Y, Wilkinson A.C, Buettner F., Nishikawa S.I., Piterman N., Kouskoff V., Theis F.J., Fisher J. and Göttgens B.
Decoding the Regulatory Network of Early Blood Development from Single-Cell Gene Expression Measurements
Nature Biotechnology 33:269–276, 2015
- Nusser-Stein S., Beyer A., Rimann I., Adamczyk M., Piterman N., Hajnal A. and Fisher J.
Cell-Cycle Regulation of Notch Signaling during C. elegans Vulval Development
Molecular Systems Biology 8:618, 2012
- Fisher J. and Henzinger T.A.
Executable Cell Biology
Nature Biotechnology 25(11):1239-1249, 2007
 Close
Close








