- Professor David Lomas

Antitrypsin deficiency and the serpinopathies: disease mechanisms and therapeutic interventions
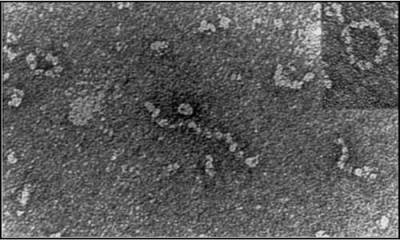
My research group has worked for over 20 years on understanding the pathogenesis of a1-antitrypsin deficiency. We were the first to show that mutant Z a1-antitrypsin forms polymers within the liver and that these comprise the PAS positive inclusions that are associated with disease.
We showed that the same process also underlies the deficiency of the Siiyama, Mmalton, S and I variants and more recently have described the a1-antitrypsin Kings mutant that also forms polymers in association with hepatic inclusions and profound plasma deficiency.
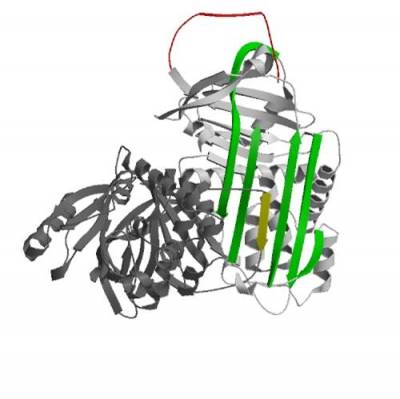
We have dissected the pathway of polymerisation using crystallography, biochemistry, biophysical analysis and have generated monoclonal antibodies to the pathological polymers associated with disease.
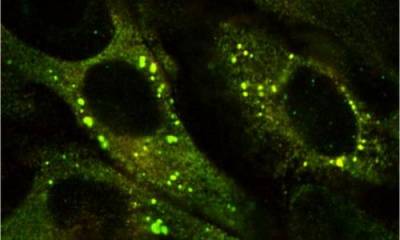
We have also probed the formation of polymers by developing cell, fly and animals models of disease. Our understanding of the process of polymerisation has allowed us to advance new paradigms for both the liver and lung disease associated with a1-antitrypsin deficiency.
The lung disease results from a process that is more complex than solely plasma deficiency. Z a1-antitrypsin is present in the lungs from passive diffusion and is also produced locally by bronchial epithelial cells and macrophages.
We have shown that this protein can spontaneously form polymers within the lung and that these polymers are chemotactic for neutrophils in vitro and in animal models of disease. Thus the lung disease is a combination of deficiency of a major protease inhibitor, inactivation of this inhibitor by the process of polymerisation and the conversion to a conformation that acts as a pro-inflammatory stimulus to recruit and retain neutrophils within the interstitium.
We have shown that the process of polymerisation also occurs in mutants of other members of the serpin superfamily in association with disease. Specifically mutants of antithrombin, C1-inhibitor and a1-antichymotrypsin form polymers that result in plasma deficiency in association with thrombosis, angioedema and emphysema respectively.
This process is most strikingly displayed in mutants of neuroserpin in association with the autosomal, inclusion body dementia that we have called familial encephalopathy with neuroserpin inclusion bodies or FENIB.
We have grouped these conditions together as a new class of disease that we have called the serpinopathies. My long-term aim is to develop strategies to block polymerisation in vitro and in vivo and so provide a cure for a1-antitrypsin deficiency and the serpinopathies.
Key Publications (2017-)
- Alpha1-Antitrypsin Deficiency. Strnad P, McElvaney NG, Lomas DA.
- N Engl J Med. 2020 Apr 9;382(15):1443-1455.
- Reduction in All-Cause Mortality with Fluticasone Furoate/Umeclidinium/Vilanterol in COPD Patients. Lipson DA, Crim C, Criner GJ, Day NC, Dransfield MT, Halpin DMG, Han MK, Jones CE, Kilbride S, Lange P, Lomas DA, Lettis S, Manchester P, Martin N, Midwinter D, Morris A, Pascoe SJ, Singh D, Wise RA, Martinez FJ; IMPACT investigators. Am J Respir Crit Care Med. 2020 Mar 12. doi: 10.1164/rccm.201911-2207OC. [Epub ahead of print]
- Blood eosinophils and treatment response with triple and dual combination therapy in chronic obstructive pulmonary disease: analysis of the IMPACT trial. Pascoe S, Barnes N, Brusselle G, Compton C, Criner GJ, Dransfield MT, Halpin DMG, Han MK, Hartley B, Lange P, Lettis S, Lipson DA, Lomas DA, Martinez FJ, Papi A, Roche N, van der Valk RJP, Wise R, Singh D. Lancet Respir Med. 2019 Sep;7(9):745-756. doi: 10.1016/S2213-2600(19)30190-0. Epub 2019 Jul 4.
- Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Sakornsakolpat P, Prokopenko D, Lamontagne M, Reeve NF, Guyatt AL, Jackson VE, Shrine N, Qiao D, Bartz TM, Kim DK, Lee MK, Latourelle JC, Li X, Morrow JD, Obeidat M, Wyss AB, Bakke P, Barr RG, Beaty TH, Belinsky SA, Brusselle GG, Crapo JD, de Jong K, DeMeo DL, Fingerlin TE, Gharib SA, Gulsvik A, Hall IP, Hokanson JE, Kim WJ, Lomas DA, London SJ, Meyers DA, O'Connor GT, Rennard SI, Schwartz DA, Sliwinski P, Sparrow D, Strachan DP, Tal-Singer R, Tesfaigzi Y, Vestbo J, Vonk JM, Yim JJ, Zhou X, Bossé Y, Manichaikul A, Lahousse L, Silverman EK, Boezen HM, Wain LV, Tobin MD, Hobbs BD, Cho MH; SpiroMeta Consortium; International COPD Genetics Consortium. Nat Genet. 2019 Mar;51(3):494-505. doi: 10.1038/s41588-018-0342-2. Epub 2019 Feb 25.
- Characterisation of a type II functionally-deficient variant of alpha-1-antitrypsin discovered in the general population. Laffranchi M, Elliston ELK, Gangemi F, Berardelli R, Lomas DA, Irving JA, Fra A.
- PLoS One. 2019 Jan 11;14(1):e0206955. doi: 10.1371/journal.pone.0206955. eCollection 2019.
- hiPSC hepatocyte model demonstrates the role of unfolded protein response and inflammatory networks in α1-antitrypsin deficiency. Segeritz CP, Rashid ST, de Brito MC, Serra MP, Ordonez A, Morell CM, Kaserman JE, Madrigal P, Hannan NRF, Gatto L, Tan L, Wilson AA, Lilley K, Marciniak SJ, Gooptu B, Lomas DA, Vallier L. J Hepatol. 2018 Oct;69(4):851-860. doi: 10.1016/j.jhep.2018.05.028. Epub 2018 Jun 5.
- α1-Antitrypsin Polymerizes in Alveolar Macrophages of Smokers With and Without α1-Antitrypsin Deficiency. Bazzan E, Tinè M, Biondini D, Benetti R, Baraldo S, Turato G, Fagiuoli S, Sonzogni A, Rigobello C, Rea F, Calabrese F, Foschino-Barbaro MP, Miranda E, Lomas DA, Saetta M, Cosio MG.
- Chest. 2018 Sep;154(3):607-616. doi: 10.1016/j.chest.2018.04.039. Epub 2018 May 12.
- Once-Daily Single-Inhaler Triple versus Dual Therapy in Patients with COPD. Lipson DA, Barnhart F, Brealey N, Brooks J, Criner GJ, Day NC, Dransfield MT, Halpin DMG, Han MK, Jones CE, Kilbride S, Lange P, Lomas DA, Martinez FJ, Singh D, Tabberer M, Wise RA, Pascoe SJ; IMPACT Investigators. N Engl J Med. 2018 May 3;378(18):1671-1680. doi: 10.1056/NEJMoa1713901. Epub 2018 Apr 18.
- Heteropolymerization of α-1-antitrypsin mutants in cell models mimicking heterozygosity. Laffranchi M, Berardelli R, Ronzoni R, Lomas DA, Fra A. Hum Mol Genet. 2018 May 15;27(10):1785-1793. doi: 10.1093/hmg/ddy090.
- The pathological Trento variant of alpha-1-antitrypsin (E75V) shows nonclassical behaviour during polymerization. Miranda E, Ferrarotti I, Berardelli R, Laffranchi M, Cerea M, Gangemi F, Haq I, Ottaviani S, Lomas DA, Irving JA, Fra A. FEBS J. 2017 Jul;284(13):2110-2126. doi: 10.1111/febs.14111. Epub 2017 Jun 8.
- FULFIL Trial: Once-Daily Triple Therapy for Patients with Chronic Obstructive Pulmonary Disease.
- Lipson DA, Barnacle H, Birk R, Brealey N, Locantore N, Lomas DA, Ludwig-Sengpiel A, Mohindra R, Tabberer M, Zhu CQ, Pascoe SJ. Am J Respir Crit Care Med. 2017 Aug 15;196(4):438-446. doi: 10.1164/rccm.201703-0449OC.
Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Hobbs BD, de Jong K, Lamontagne M, Bossé Y, Shrine N, Artigas MS, Wain LV, Hall IP, Jackson VE, Wyss AB, London SJ, North KE, Franceschini N, Strachan DP, Beaty TH, Hokanson JE, Crapo JD, Castaldi PJ, Chase RP, Bartz TM, Heckbert SR, Psaty BM, Gharib SA, Zanen P, Lammers JW, Oudkerk M, Groen HJ, Locantore N, Tal-Singer R, Rennard SI, Vestbo J, Timens W, Paré PD, Latourelle JC, Dupuis J, O'Connor GT, Wilk JB, Kim WJ, Lee MK, Oh YM, Vonk JM, de Koning HJ, Leng S, Belinsky SA, Tesfaigzi Y, Manichaikul A, Wang XQ, Rich SS, Barr RG, Sparrow D, Litonjua AA, Bakke P, Gulsvik A, Lahousse L, Brusselle GG, Stricker BH, Uitterlinden AG, Ampleford EJ, Bleecker ER, Woodruff PG, Meyers DA, Qiao D, Lomas DA, Yim JJ, Kim DK, Hawrylkiewicz I, Sliwinski P, Hardin M, Fingerlin TE, Schwartz DA, Postma DS, MacNee W, Tobin MD, Silverman EK, Boezen HM, Cho MH; COPDGene Investigators; ECLIPSE Investigators; LifeLines Investigators; SPIROMICS Research Group; International COPD Genetics Network Investigators; UK BiLEVE Investigators; International COPD Genetics Consortium. Nat Genet. 2017 Mar;49(3):426-432. doi: 10.1038/ng.3752. Epub 2017 Feb 6.
Group members
- James Irving
- Riccardo Ronzoni
- Alistair Jagger
- Ibrahim Aldobiyan
- Kamila Kamuda
- Narinder Heyer-Chauhan
- Professor David Sattelle

Molecular Neurobiology
We are interested in the mechanisms underlying neurodegenerative and neuromuscular diseases and in exploring new routes to therapy. We address these goals using the powerful genetic model organism Caenorhabditis elegans with its transgenic lines, mutants as well as access to forward and reverse genetics. Current interests include Alzheimer's disease, Fronto-Temporal Dementia, Spinal Muscular Atrophy and Congenital Myasthenia. We have developed high-throughput, automated, phenotyping, which permits library-scale, in vivo, drug screening of our disease models with the aim of finding new or re-profiled drugs to ameliorate symptoms of these devastating diseases.
We also study human ligand-gated ion channels as targets for drugs aimed at ameliorating symptoms of nervous system disorders. Invertebrate ligand-gated ion channels are investigated as targets for drugs / chemicals with benefits for animal health and agriculture.Selected publications:
- Sleigh J.N., Buckingham S.D., Esmaeili, B.E., Viswanathan M., Westlund B.M. and Sattelle, D.B. (2011) A novel Caenorhabditis elegans allele smn-1(cb131), mimicking a mild form of spinal muscular atrophy, provides a convenient drug screening platform highlighting new and pre-approved compounds. Hum Mol Genet 20, 245-260.
- Werren J.H. et al. (2010) Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327, 343-348.
- Briese, M., Esmaeili, B., Fraboulet, S., Burt, E., Christodoulou, S.C., Towers, P.R., Davies, K.E. and Sattelle, D.B. (2009). Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum Mol Genet. 18, 97-104.
- Jones A.K., Buckingham S.D., Papadaki M., Yokota M., Sattelle B.M., Matsuda K. and Sattelle D.B. (2009) Splice-variant- and stage-specific RNA editing of the Drosophila GABA receptor modulates agonist potency. J Neuroscience 29, 4287-4292.
- Richards, S. et al (2008) The genome of the model beetle and pest Tribolium castaneum (2008) Nature 452, 949- 955.
- Weinstock G. et al (2006) Insights into social insects from the genome of the honeybee Apis mellifera Nature 443: 931-949.
- Jones, A.K., Buckingham, S.D., and Sattelle, D.B (2005) Chemistry-to-gene screens in Caenorhabditis elegans. Nature Rev. Drug Discov. 4, 321-330.
Group members
Dr Freddie Partridge
Mrs Marina Nick
Dr Narinder Heyer-Chauhan
 Close
Close

