New publication in Nature for Baum and Henriques labs
26 August 2020
Closed mitosis requires local disassembly of the nuclear envelope.
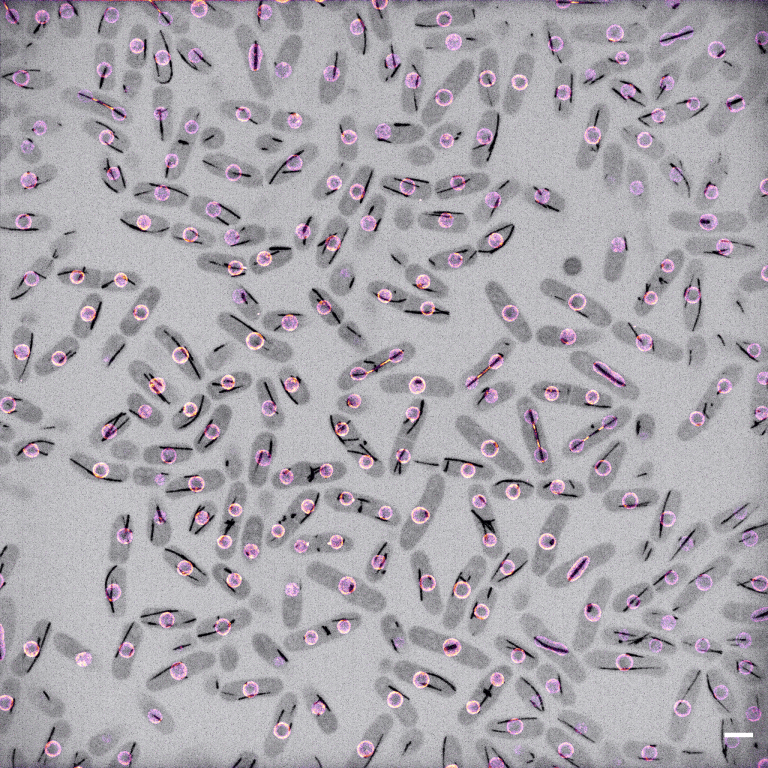
As eukaryotic cells divide, the nucleus must be remodelled into two distinct compartments, each encapsulating a single copy of the genome. Eukaryotes have evolved a bewildering range of strategies to achieve this - at one extreme, by disassembling the entire structure as cells begin to divide and reassembling it once division is almost complete. This strategy, employed by most animal and plant cells, is called an ‘open' mitosis. At the other extreme, most fungi and many protists divide an intact nucleus in lockstep with cell division in a ‘closed’ mitosis.
How the nucleus can be divided in a closed mitosis without a loss of integrity has long remained a mystery. In their recent publication in Nature, Dey and colleagues show that the fission yeast Schizosaccharomyces pombe divides its nucleus by isolating and selectively disassembling a subset of nuclear pore complexes (NPCs) in the narrow bridge that links daughter nuclei. They identify an uncharacterised protein, Les1, that is responsible for corralling NPCs within the bridge midzone and for insulating daughter nuclei from the disassembly process.
The process of local nuclear envelope disassembly identified by the authors shares key features with the disassembly of the nuclear envelope in open mitosis, suggesting for the first time that universal mechanisms might unite diverse mitotic strategies in eukaryotes once thought to be completely distinct.
Links:
- Research paper in Nature
- Professor Buzz Baum's academic profile
- Dr Ricardo Henriques' academic profile
- UCL MRC Laboratory for Molecular Cell Biology
- UCL Institute for the Physics of Living Systems
Image
- Schizosaccharomyces pombe cells dividing. Les1 in colour, tubulin in inverted grayscale. Scale bar 5 microns.
 Close
Close

