MedICSS includes interactive sessions throughout the week, including group mini projects.
- Deep Learning for Neurodegenerative Disease Classification using Multimodality MRI Imaging
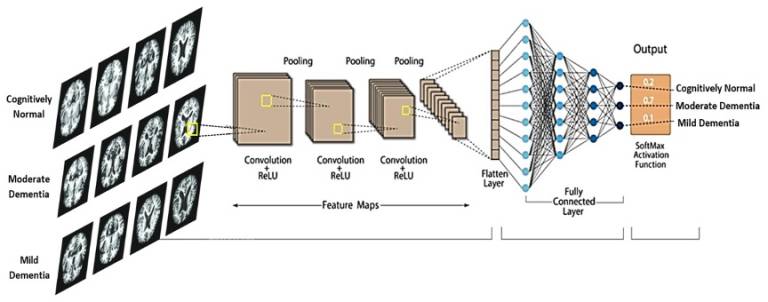
Lead person: Moona Mazher (Postdoc Research Fellow)
Co-leads: Elinor Thompson (Postdoc Research Fellow)
Description: Nowadays, one of the most successful modern deep-learning applications in medical imaging is image classification. Disease classification in medical imaging is of significant importance due to its potential impact on patient care, diagnosis, treatment planning, and overall healthcare delivery. Early detection of diseases is often critical for effective treatment and improved patient outcomes. Medical imaging techniques, such as MRI, allow healthcare professionals to identify abnormalities at an early stage, leading to timely interventions. Similarly, accurate disease classification aids in formulating appropriate treatment plans.
Neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease affect millions of people worldwide. The classification of these diseases is crucial for several reasons, as it helps in understanding, diagnosing, and treating these complex disorders. Accurate classification enables healthcare professionals to make precise diagnoses based on specific symptoms, biomarkers, and imaging findings. This understanding is crucial for developing targeted and effective therapeutic interventions, as different diseases may require distinct treatment approaches.
This tutorial project will guide students to build and train a state-of-the-art convolutional neural network from scratch, applying it to real brain MRI scans to classify them according to disease categories.
The objective of this project is to obtain.
Understanding of different structural MRI modalities.
A comprehensive knowledge sharing on neurodegeneration and its subtypes.
Basic understanding of how to read volumetric MRI data as well as 2D images for classification.
A basic understanding of deep learning approaches applied for medical image classification.
Obtain hands-on experience building of state-of-the-art deep learning model for image classification from scratch.
Prerequisites: Python, GPU (via Colaboratory)
- Image Quality Transfer
Lead persons: Matteo Figini, Henry Tregidgo, Alp Cicimen
Description: Image Quality Transfer (IQT) is a machine learning based framework to propagate information from state-of-the-art imaging systems into a clinical environment where the same image quality cannot be achieved [1]. It has been successfully applied to increase the spatial resolution of diffusion MRI data [1, 2] and to enhance both contrast and resolution in images from low-field scanners [3]. In this project, we will explore the deep learning implementation of IQT and investigate the effect of the different parameters and options, using MRI data from publicly available MRI databases. We will test the algorithms on simulations and on real clinical data.
[1] D. Alexander et al., “Image quality transfer and applications in diffusion MRI”, Neuroimage 2017. 152:283-298
[2] R. Tanno, et al. “Uncertainty modelling in deep learning for safer neuroimage enhancement: Demonstration in diffusion MRI.” NeuroImage 2021. 225
[3] M. Figini et al., “Image Quality Transfer Enhances Contrast and Resolution of Low-Field Brain MRI in African Paediatric Epilepsy Patients”, ICLR 2020 workshop on Artificial Intelligence for Affordable Healthcare
Further details: The code we will be using is written in TensorFlow. Previous experience with TensorFlow, deep learning and/or brain MRI is desired but not strictly necessary.
- Implementing Reproducible Medical Image Analysis Pipelines
Lead person: Dave Cash
Co-leads: Haroon Chughtai
Description: This project provides a demonstration of how to implement a reproducible medical image analysis pipeline for scalable, high throughput analysis without the need for substantial experience of coding. It will also show how solutions for maintaining the privacy of study participants can be implemented with low overhead. It will use all open source software, in particular the data and analysis will be managed through XNAT, a widely used web-based platform. In this tutorial, we will go through the benefits this platform for automating handling, importing, and cleaning of DICOM data, conversion to Nifti, de-facing structural T1 data to provide additional assurance of privacy, and finally volumetric and cortical thickness analysis using FastSurfer, a free deep learning implementation of FreeSurfer. The project will determine how much de-facing algorithms change the results of FastSurfer compared to the original images.
Further courses and tutorials on medical image data management, software development, and medical image analysis will soon be available at Health and Bioscience IDEAS, a UKRI-funded training program around medical imaging for UK researchers.
- Deep Learning for Medical Image Segmentation and Registration
Lead person: Yipeng HuDescription: One of the most successful modern deep-learning applications in medical imaging is image segmentation. From neurological pathology in MR volumes to fetal anatomy in ultrasound videos, from cellular structures in microscopic images to multiple organs in whole-body CT scans, the list is ever expanding.
This tutorial project will guide students to build and train a state-of-the-art convolutional neural network from scratch, then validate it on real patient data.The objective of this project is to obtain
- Basic understanding of machine learning approaches applied for medical image segmentation,
- Practical knowledge of essential components in building and testing deep learning algorithms, and
- Obtain hands-on experience in coding a deep segmentation network for real-world clinical applications.
Prerequisites: Python, GPU (via Colaboratory)
- De-Noising Magnetic Resonance Images with Deep Learning
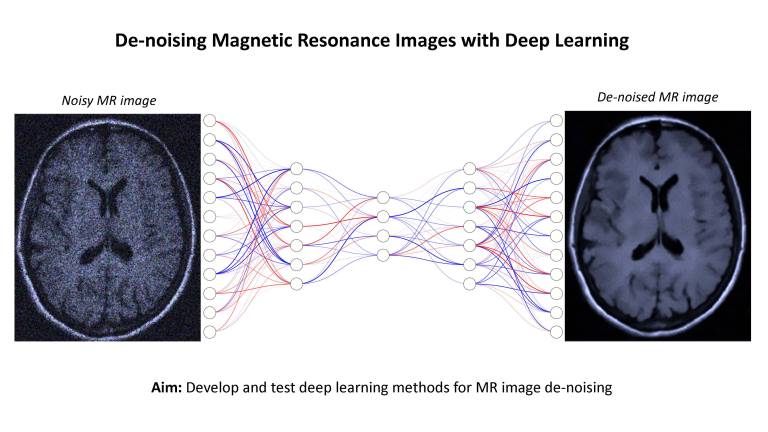
Lead person: Christopher Parker
Description: Magnetic resonance (MR) images contain unique anatomical and physiological information that is used to guide patient healthcare and inform research studies. However, MR images are contaminated by random noise, introducing uncertainty and errors in their downstream applications. Reducing noise may enable more effective use of MR imaging in scenarios that would otherwise require longer scan times or higher field strengths, with potential to increase accessibility.
This project aims to test deep learning-based methods for MR image de-noising, building on CMIC’s technological advances in deep neural networks and MR image modelling1,2,3. During the project students will learn about MR imaging, noise contamination, deep learning, and quantitative MR imaging. Experience in Python programming and a laptop is required. Experience in PyTorch and image processing would be useful but is not essential.
Recommended reading:
[1] Parker, C.S., Schroder, A., Epstein, S.C., Cole, J., Alexander, D.C. and Zhang, H., 2023. Rician likelihood loss for quantitative MRI using self-supervised deep learning. arXiv preprint arXiv:2307.07072.
[2] Epstein, S.C., Bray, T.J., Hall-Craggs, M. and Zhang, H., 2022. Choice of training label matters: how to best use deep learning for quantitative MRI parameter estimation. arXiv preprint arXiv:2205.05587.
[3] Alexander, D.C., Dyrby, T.B., Nilsson, M. and Zhang, H., 2019. Imaging brain microstructure with diffusion MRI: practicality and applications. NMR in Biomedicine, 32(4), p.e3841.- Automated Error Detection in Robot-Assisted Surgery
- Lead person: Jialang Xu
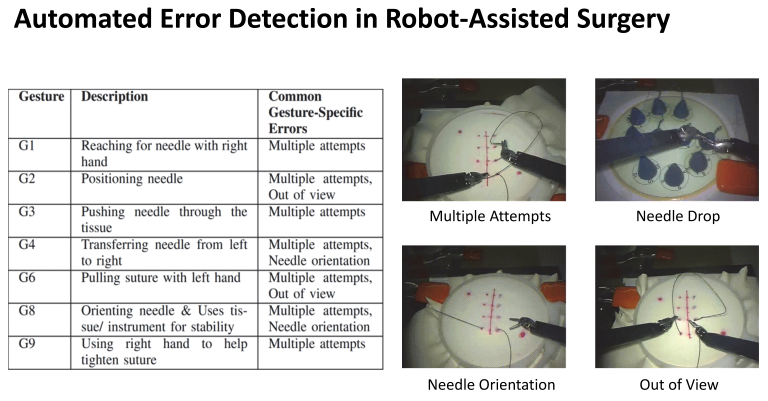
Co-leads: Evangelos Mazonmenos
Description: Detecting errors from surgical data is a critical task in advancing automated surgical skill assessment, with the potential to significantly enhance the safety and efficiency of surgical training and procedures through the delivery of data-driven feedback to surgeons. This tutorial project aims to detect executional errors (e.g. multiple attempts and out of view) by analyzing the time-series visual features extracted from robot-assisted surgical videos. The dataset utilized in this project was captured by the da Vinci Surgical System from eight surgeons with different levels of skill on a bench-top model. This project will provide participants with hands-on experience in building and training state-of-the-art deep learning methods for surgical error detection tasks.
References:
[1] Li, Zongyu, Kay Hutchinson, and Homa Alemzadeh. "Runtime detection of executional errors in robot-assisted surgery." 2022 International conference on robotics and automation (ICRA). IEEE, 2022.
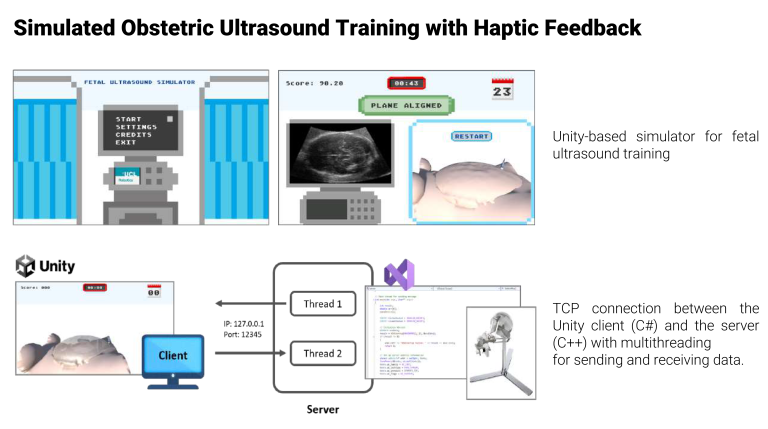
Pre-requisites: GPU, laptop, Python, Pytorch. - Simulated Obstetric Ultrasound Training with Haptic Feedback
- Lead person: Chiara Di Vece
Co-leads: Prof. Francisco Vasconcelos
Description: Fetal ultrasound is a crucial non-invasive imaging tool used to monitor fetal growth, detect abnormalities, and assess biometry. The mid-trimester scan, typically performed between 18 to 22 weeks of gestation, is essential for evaluating fetal development. Accurate acquisition of standard planes (SP), such as the transventricular view of the fetal head, is crucial for assessing fetal biometry and detecting growth disorders.
Novice sonographers often face challenges in acquiring high-quality SPs due to the complex fetal anatomical structure and the need for precise transducer manipulation. Traditional training methods, such as bedside teaching, may be insufficient for building the necessary skills, and simulation-based learning has proven effective in providing a safe and controlled environment for practice.
The project builds on previous research that aimed at developing a realistic simulation for training novice sonographers in performing mid-trimester obstetric ultrasound scans. The simulation utilizes a three-dimensional game-type environment and incorporates a robotic haptic device (Sigma.7) to provide tactile feedback, mimicking the physical interaction of a probe sliding on the abdomen.
Aims and Objectives:
- Scoring System: Implement a scoring system to assess the alignment of acquired SPs with predefined anatomical planes in real time, providing quantitative feedback to trainees.
- Haptic Feedback Integration: Integrate the Sigma.7 robotic haptic device to simulate the physical sensations of probe manipulation, enhancing the realism of the training experience via TCP server-client communication protocols.
Technical Requirements:
- Familiarity with C# programming language for scripting within Unity;
- Familiarity with C++ programming language for creating the communication between the simulator and the haptic device.
Software/Hardware Requirements:
- The project will be developed at Charles Bell House (UCL), where the haptic device is located;
- The participants will be provided with a laptop with the simulation environment and all the drivers already installed.
- DARES: Depth Anything in Robotic Endoscopic Surgery with Low-Ranked Adaptation of the Foundation Model
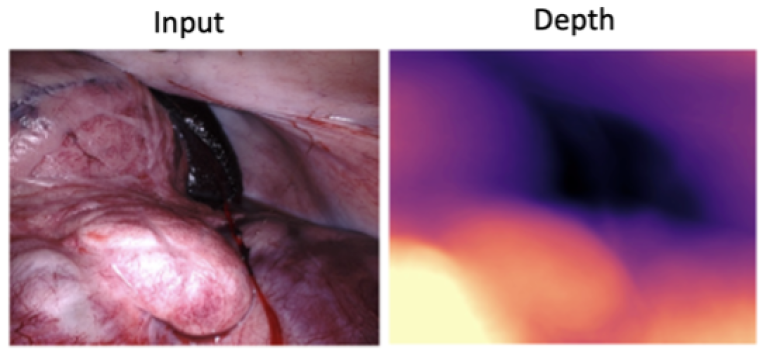
Lead person: Mobarakol Islam
Description: The project emerges from the critical need to enhance depth perception in robotic surgery. This need is underscored by the role of depth estimation in facilitating 3D reconstruction, surgical navigation, and augmented reality visualization, which are pivotal for the success and safety of surgical procedures. Despite the foundation model's remarkable capabilities in a broad spectrum of vision tasks, including depth estimation, its application within the medical and surgical domains has encountered substantial challenges. These challenges are primarily due to the model's limitations when dealing with domain-specific nuances in medical imagery. Addressing this gap, the project introduces a low-ranked adaptation (LoRA) of the foundation models (DINO V2 [1,2] or Depth Anything [3]), tailored for the intricate requirements of surgical depth estimation. This adaptation aims to offer a robust solution for monocular depth estimation, overcoming the prevalent limitations by focusing on a scalable dataset approach in surgery.
Project Aims
- Enhancing the foundation model through a low-ranked adaptation (LoRA) that is specifically optimized for the complexities of surgical imagery.
- Scaling up the dataset to improve the model's versatility and its ability to generalize across a wide range of surgical scenarios [4].
- Providing a practical, adaptable solution that can be seamlessly integrated into robotic surgery workflows, thereby improving surgical precision and patient outcomes.
Key Learning Outcomes
- Gain deep insights into depth estimation in endoscopic surgery and its monocular challenges.
- Learn to fine-tune foundation models for surgical applications via the LoRA technique.
- Acquire expertise in scaling and managing datasets to boost model performance.
- Encourage innovation and creative solutions in surgical technology development.
Requirements
- Basic understanding of the depth estimation
- Hands on experience of using a deep learning framework (Pytorch/Tensorflow)
References
[1] Cui, B., Islam, M., Bai, L., Ren, H. "Surgical-DINO: Adapter Learning of Foundation Model for Depth Estimation in Endoscopic Surgery." IPCAI 2024.
[2] Oquab, M., et al. "Dinov2: Learning robust visual features without supervision." arXiv preprint arXiv:2304.07193 (2023).
[3] Yang, Lihe, et al. "Depth anything: Unleashing the power of large-scale unlabeled data." arXiv preprint arXiv:2401.10891 (2024).
[4] Allan, Max, et al. "Stereo correspondence and reconstruction of endoscopic data challenge." arXiv preprint arXiv:2101.01133 (2021).
- Aortic 3D Deformation Reconstruction using 2D X-ray Fluoroscopy and 3D Pre-operative Data for Endovascular Interventions
Lead person: Shuai Zhang
Co-leads: Evangelos Mazonmenos
Description: Current clinical endovascular interventions rely on 2D guidance for catheter manipulation. Although an aortic 3D surface is available from the pre-operative CT/MRI imaging, it cannot be used directly as a 3D intra-operative guidance since the vessel will deform during the procedure. This tutorial aims to reconstruct the live 3D aortic deformation by fusing the static 3D model from the pre-operative data and the 2D live imaging from fluoroscopy. The dataset used in this project was captured by a Silicone aortic phantom. This project will provide participants with hands-on experience in formulating and solving a non-linear optimization-based 2D-3D registration problem based on the deformation graph approach.
- Tractography: modelling connections in the human brain
Leaders: Lawrence Binding and Anna Schroder
Description: Tractography [1] is currently the only tool available to probe non-invasively the structural connectivity of the brain in-vivo. However, tractography is subject to extensive modelling errors, causing a large number of false positive and false negative connections in the resulting connectivity matrix [2]. Despite these drawbacks, tractography has revolutionised our understanding of the brain’s connectivity architecture over the past few decades, and has wide-spread potential applications, from surgical planning [3] to modelling the spread of neurodegenerative diseases through the brain [4].
This project will introduce participants to the basic principles of tractography. Participants will have the opportunity to implement tractography algorithms from scratch, and compare these results to state-of-the-art tractography software tools in MRtrix3 [5].
References:
[1] Jeurissen, B., et al. (2019). Diffusion MRI fiber tractography of the brain. NMR in Biomedicine, 32(4), p.e3785.
[2] Maier-Hein, K.H., et al. (2017). The challenge of mapping the human connectome based on diffusion tractography. Nature communications, 8(1), pp.1-13.
[3] Walid E.I. et al (2017). White matter tractography for neurosurgical planning: A topography-based review of the current state of the art. NeuroImage: Clinical. 15 pp659-672
[4] Vogel, J., et al. (2020). Spread of pathological tau proteins through communicating neurons in human Alzheimer’s disease. Nature Communications 11, 2612
[5] Tournier, J.D., et al. (2019). MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage, 202, p.116137.
Prerequisites:
– Knowledge: MATLAB or python
– Equipment: Own laptop, no GPU needed
 Close
Close

