Using the world’s most powerful supercomputers to tackle COVID-19
7 April 2020
The world’s most powerful supercomputers are being used by UCL researchers for urgent investigations into the SARS-CoV-2 virus and the associated COVID-19 disease with the aim of accelerating the development of treatments, including antiviral drugs and vaccines.

Professor Peter Coveney, who leads the EU H2020 Computational Biomedicine Centre of Excellence, and his colleagues at the UCL Centre for Computational Science, are part of a consortium of more than a hundred researchers from across the US and Europe, who are using an exceptional array of supercomputers – including the biggest one in Europe and the most powerful on the planet – to study several aspects of the virus and disease in detail.
The concerted research effort focuses on five areas:
- identifying new antiviral drugs by screening libraries of potential drugs, including those that have already been licensed to treat other diseases
- accelerating vaccine development by identifying virus proteins or parts of protein that stimulate immunity
- studying the spread of the virus within communities
- analysing the origin and structure of the SARS-CoV-2 genome
- studying how the SARS-CoV-2 virus interacts with human cells to turn them into virus factories
The UCL team has access to supercomputers at Argonne Leadership Computing Facility, Brookhaven, the Texas Advanced Computing Center, Oak Ridge, the San Diego Supercomputing Center, the Gauss Centre for Supercomputing (GCS) at Leibniz Rechenzentrum (LRZ) and the Hartree Centre.
They are using two of the world’s most powerful supercomputers – Summit at Oak Ridge National Lab, USA (1st) and SuperMUC-NG at GCS@LRZ, Germany (9th) – to screen libraries of drug compounds to identify those capable of binding to the spikes on the surface of the novel coronavirus, which the virus uses to invade cells, so as to prevent it from infecting human cells.
The libraries include known and licensed drugs for quick repurposing opportunities (e.g. DrugBank), 100 million known small molecules that are drug like (e.g. PubChem) and large-scale libraries (e.g. Enamine, ZINC) with billions of compounds that could be manufactured quickly for testing.
Professor Coveney (UCL Chemistry), said: “We are using the immense power of supercomputers to rapidly search vast numbers of potential compounds that could inhibit the novel coronavirus, and using the same computers again, but with different algorithms, to refine that list to the compounds with the best binding affinity. That way, we are identifying the most promising compounds ahead of further investigations in a traditional laboratory to find the most effective treatment or vaccination for COVID-19.
“We are able to scan existing drug libraries, so many of the compounds we are looking at already have approval for use in humans as they are used to treat existing diseases so could be repositioned to target COVID-19. We are also able to computer generate new compounds that should bind well to the virus, which gives us a fantastic head start on discovering potential new drugs.”
The compounds screened include chemicals, herbal medicines, and natural products that have either been studied in humans or are already approved drugs and therefore are already considered safe for humans. The supercomputers are able to complete the scanning tasks in days, where it would take regular computers months.
“This is a much quicker way of finding suitable treatments than the typical drug development process. It normally takes pharma companies 12 years and $2 billion to take one drug from discovery to market but we are rewriting the rules by using powerful computers to find a needle in a haystack in a fraction of that time and cost,” he added.
“Supercomputers are a remarkable resource for the development of COVID-19 treatments as they can identify possible treatments through a variety of ways including machine learning, complex molecular dynamics and artificial intelligence methods. Not only do we need to find molecules that bind to the spikes on the coronavirus, but we also need to model how well these bind when we know the spikes move around.”
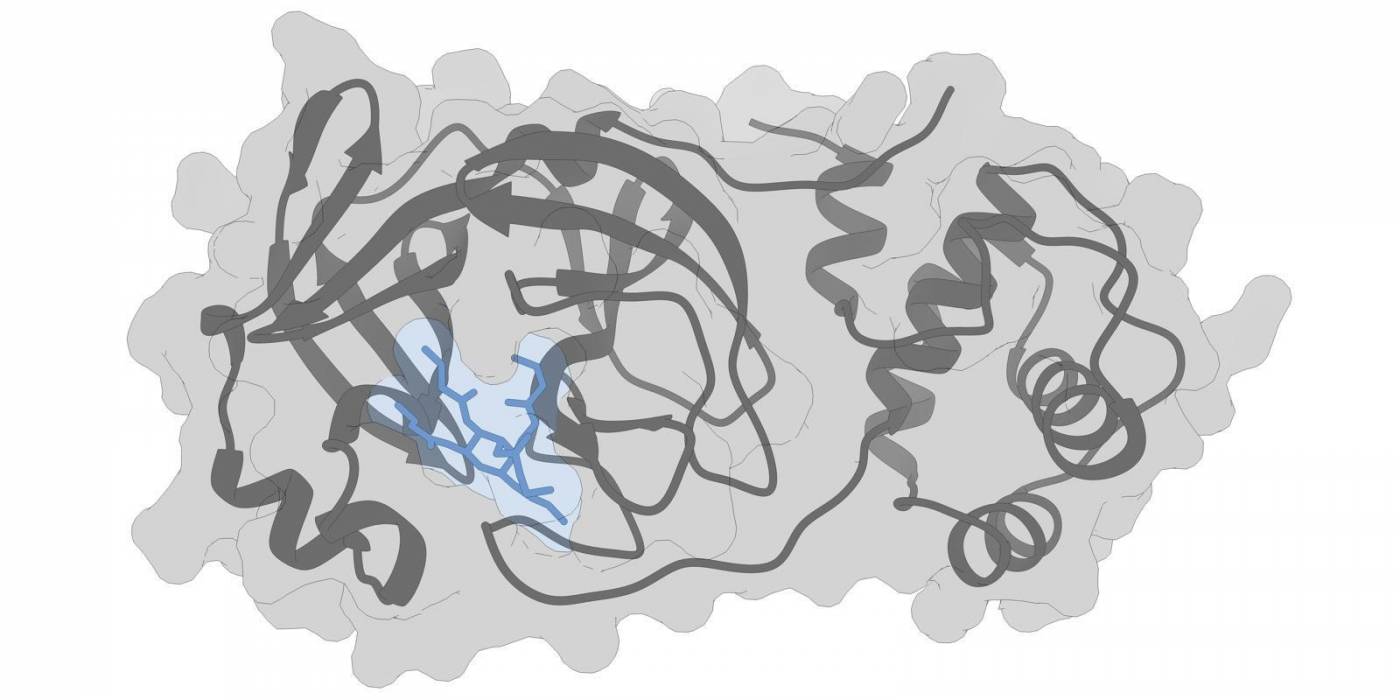
In addition to the spike proteins on the virus surface, Professor Coveney’s group is simulating how pharmaceutical drugs interact with proteins involved in various other stages of the virus lifecycle. For instance, 3CL-protease is a protein that is key to the virus replicating itself, allowing them to grow in number and cause further damage in the body. Drugs that bind well to this protein may slow or halt its replication. Simulating this drug-protein interaction can identify further drug targets.
Professor Coveney, who is leading the European side of the global effort, welcomes colleagues from across UCL to join the collaboration, and in particular, those with expertise in machine learning and generative methods for compound discovery, and physics-based methods for calculating binding free energies. Anyone with interests across the wider scope of this collaboration should also feel free to get in touch.
The consortium includes five US national laboratories (Argonne, Brookhaven, Los Alamos, Oak Ridge National Laboratory, Lawrence Livermore National Laboratory) led by Rick Stevens, nine universities (UCL, University of Chicago, University of Illinois, University of Virginia, Rutgers University, Stony Brook University, George Mason University, University of Texas, and University of California San Diego), a private research centre (JC Venter Institute), and a public academy (Leibniz Rechenzentrum of the Bavarian Academy of Sciences and Humanities).
GCS@LRZ’s role in the collaboration, headed by Director Professor Dieter Kranzlmüller, has been endorsed by the German government and GCS has already provided major supercomputing resources for the COVID-19 work, including an initial allocation of 10 million core hours on SuperMUC-NG at LRZ.
The Hartree Centre, part of STFC at Daresbury Laboratory, has committed to providing resources from their supercomputer, Scafell Pike, for an initial period of six months.
The US Advanced Photon Source and the UK Diamond Light Source – intense sources of X-rays that can be used to study molecular structures – have agreed to confirm supercomputer predictions, for instance of drugs that can latch on to and block virus proteins.
Links
Images
Summit supercomputer. Photo by Argonne National Laboratory. Summit is a next-generation IBM/NVIDIA supercomputer with an aggregate peak compute speed of over 200 PFLOP/S.
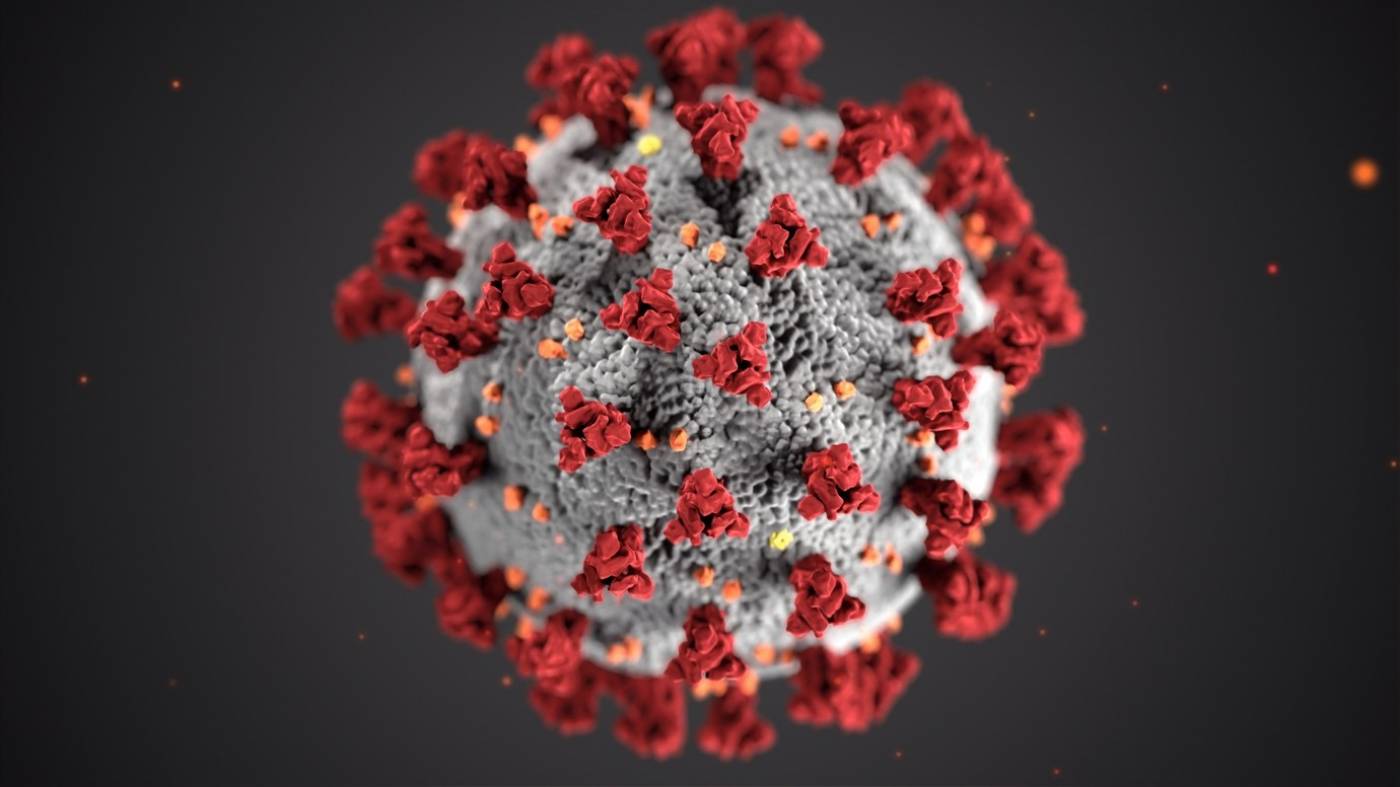
SARS-CoV-2 virus, including the spike proteins (red) around it’s surface that are targets for therapeutic drugs, and therefore candidates to be simulated interacting with drug compounds.
3CL-protease (grey), a protein in the COVID-19 disease process, and a potential repurposed pharmaceutical drug (blue) binding to the protein in a simulation model.
Media contact
Bex Caygill
Tel: +44 (0)20 3108 3846
Email: r.caygill [at] ucl.ac.uk
 Close
Close

