Group Leader: Dr Nischalan Pillay
Sarcoma Genomics
In comparison to many other tumour types, the molecular classification of sarcoma is relatively mature. There are more than 50 subtypes of sarcoma with many more benign mimickers, the vast majority of which are defined by molecular events such as gene amplifications (e.g. MDM2 amplification in dedifferentiated liposarcoma), chimeric fusion genes (e.g. EWSR1-FLI1 fusion in Ewing’ sarcoma) and point mutations (e.g. KIT in gastrointestinal stromal tumours). Whilst this molecular classification is invaluable for diagnosis the mechanism of action of the various genetic events is poorly understood. In part this has contributed to the slow progress in development of new treatments and survival rates for sarcoma patients.
Also there are still a proportion of sarcoma types where the molecular underpinnings are yet to be clearly defined. In such cases there is a lack of objective diagnostic criteria upon which to classify these tumours resulting in a lack of treatment options.
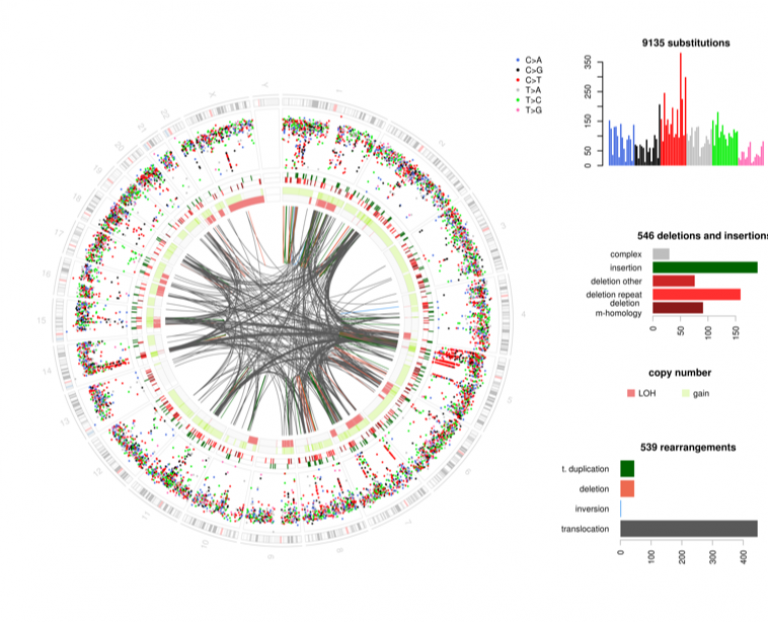
Circos plot demonstrating the wide repertoire of genome complexity in a high grade sarcoma.
Genomic characterisation of undifferentiated sarcoma
Funded by Cancer Research UK
One such tumour is undifferentiated pleomorphic sarcoma (UPS), which is the current focus of the lab. In order to address the unmet need to develop a systematic molecular classification of UPS to stratify these patients for suitable clinical trials we undertake comprehensive genomic profiling complemented by other techniques such as methylation and gene expression analysis using patient samples. The material for study is of high quality from an established biobank established by Prof. Adrienne Flanagan and supported by rich pathological annotation by clinical colleagues at the Royal National Orthopaedic Hospital NHS Trust. This work is led by Dr Christopher Steele and is being conducted in collaboration with the Wellcome Trust Sanger Institute and The Francis Crick Institute and is supported by generous funding from Cancer Research UK.
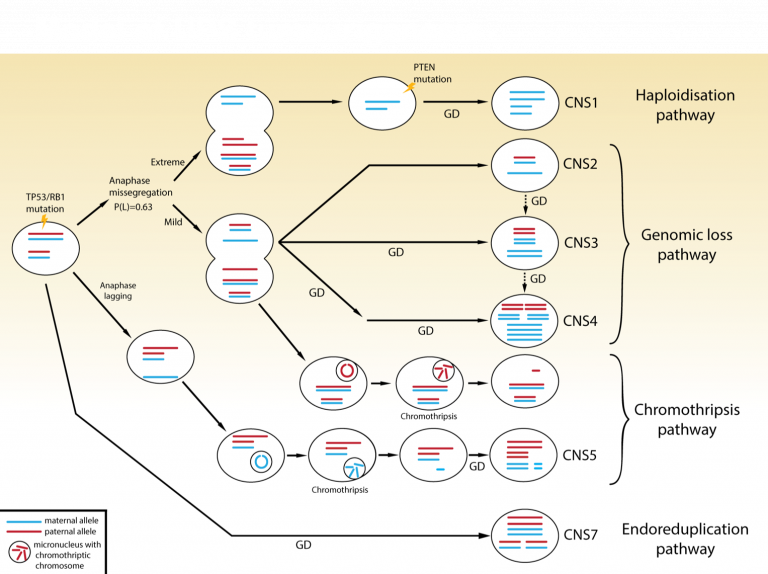
Models of undifferentiated sarcoma tumourigenesis
Digital pathology imaging of sarcomas for clinical benefit
In parallel with the work above we have an exciting new collaboration with Dr Kevin Bryson (UCL Computer Science) in order to better understand the genotype-phenotype correlations and the cellular spatial dynamics of sarcomas. This is accomplished by applying conventional learning machine techniques as well as newer deep learning methods on digitised histological images to unravel the unique architectural and cytological nuances of these enigmatic tumours. Our first project is currently being undertaken by a PhD student Dr Binghao Chai.
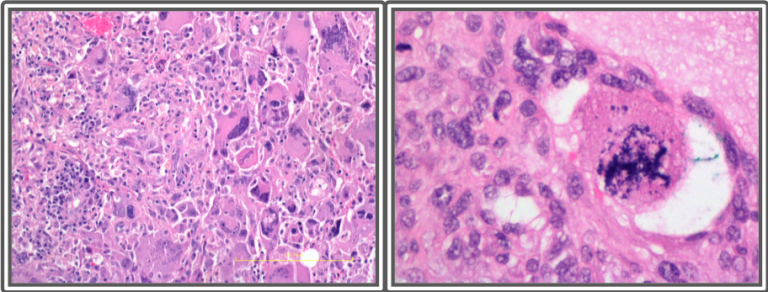
Left: Undifferentiated sarcoma with severely pleomorphic cells. Right: Atypical mitotic figures are common in undifferentiated sarcomas.
Molecular signatures in sarcoma
Funded by Sarcoma UK
Mutational signatures are emerging biomarkers for prognosis and prediction in multiple cancer types. In sarcomas with complex genomes the predominant mutational repertoire consists of large scale rearrangements and copy number alterations. This group of patients have a poor prognosis with limited treatment options. With sarcoma patients being enrolled on the 100,000 genomes project there is an opportunity to investigate their genomes for mutational signatures that could be predict which patients could benefit from immune modulation strategies and for prognostic utility. However, this evidence still needs to be generated in sarcomas. This exciting new project leverages a copy number signature framework developed in our lab by Dr. Christopher Steele and brings together leaders in the field of sarcoma biology (Prof Adrienne Flanagan - UCL ), genomics (Dr Peter Van Loo - Francis Crick Institute, Dr Sam Behjati - Wellcome Trust Sanger Institute and Dr Javier Herrero - UCL) and mutational signatures (Dr Ludmil Alexandrov – University of California San Diego). The aim of this work is to optimise rearrangement and copy number signature mathematical models and implement mutational signature frameworks in sarcoma for clinical benefit.
Methylation tumour heterogeneity in sarcoma
Our sarcoma genomic studies and work from other cancer types has shown that genomic changes such as mutations, copy number alterations, and genomic rearrangements fuel tumour heterogeneity and cancer evolution. Increasingly it is being shown that there is a dynamic inteplay between these genomic aberrations and changes in DNA methylation. In tumours such as undifferentiated pleomorphic sarcomas which are dominated by copy number changes and structural rearrangements it is still unknown how these genomic changes impact on methylation or vice versa and ultimately how this plays itself out in terms of cancer evolution.
We aim to address this work using the innovative techniques in genome and DNA methylation sequencing using a unique resource of high quality undifferentiated sarcoma samples. This work is being undertaken in collaboration with Dr. Andy Feber (UCL) and Dr. Peter Van Loo (Francis Crick Institute) and is funded by CRUK.
Last updated March 2019
 Close
Close




