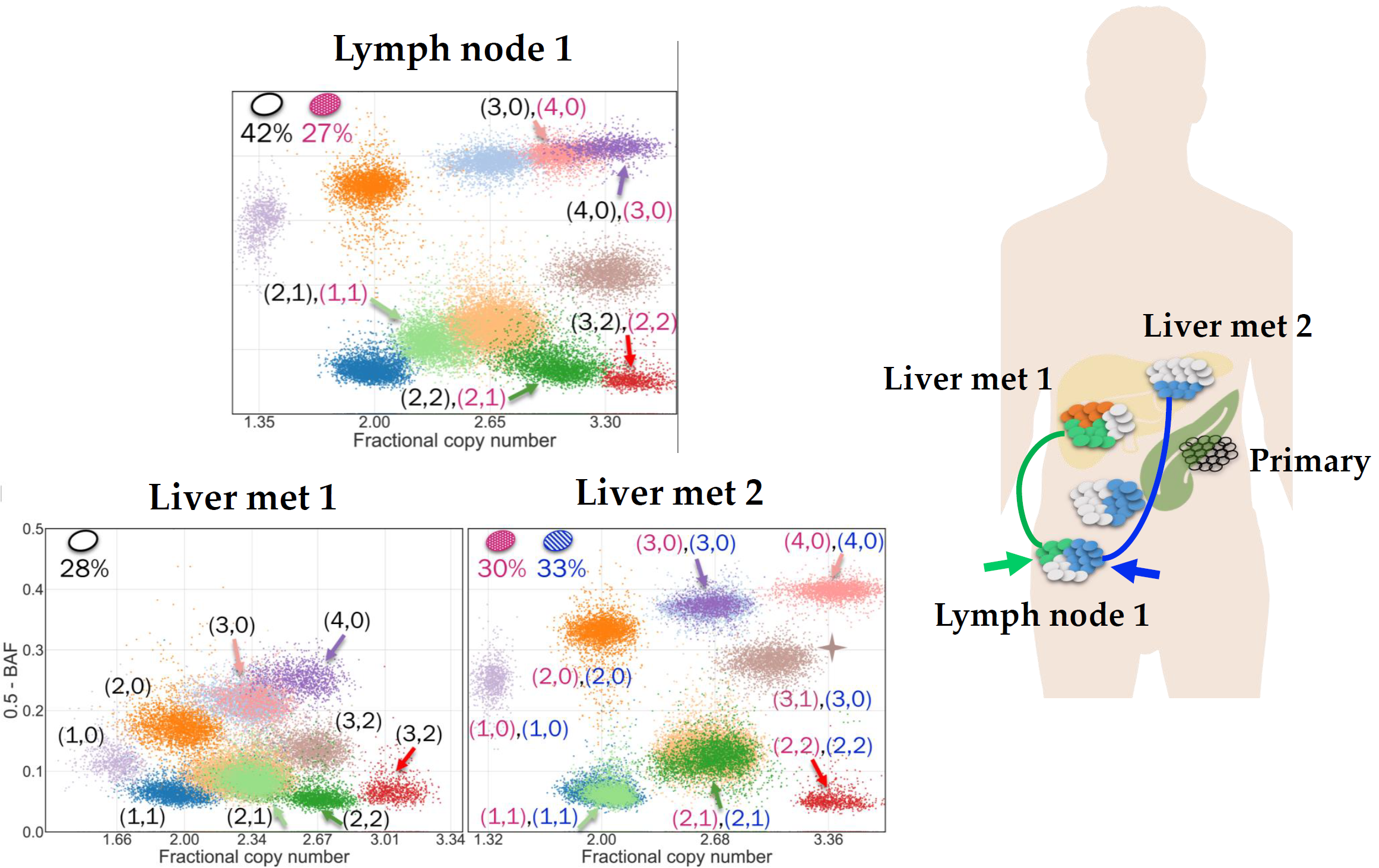
We analyse single-cell sequencing data to investigate tumour evolution from individual cells.
Metastasis results from the dissemination of cancer cells from a primary tumour to other anatomical sites and is the most common cause of death for lung cancer patients. Thus, elucidating genetic changes which engender metastatic potential could have a critical clinical impact. Despite being the primary cause of death for cancer patients, the genomic evolutionary history of metastasis and the genetic causes of this process are poorly understood.
Pioneering recent studies have demonstrated that multi-site DNA sequencing allows us to investigate the metastatic migration process. In particular, such data have been used to evaluate the presence of monoclonal vs polyclonal migrations, and to identify candidate driver mutations for metastasis. However, previous analyses have been affected by some major limitations which may hinder additional mechanisms of metastatic progression and limit their applicability.
Our lab focuses on the design and development of computational methods to analyse and reconstruct metastatic migrations patterns. Specifically, we design approaches to leverage and integrate multiple sources of information from different data and different kinds of somatic mutations.
- Relevant publications
- Zaccaria, S., Raphael, B.J. Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data. Nature Communications 11, 4301 (2020). https://doi.org/10.1038/s41467-020-17967-y
- Satas, G., Zaccaria, S., Mon, G., & Raphael, B. J. SCARLET: Single-Cell Tumor Phylogeny Inference with Copy-Number Constrained Mutation Losses. Cell Systems, 10(4): 323-332 (2020). Accepted and presented at RECOMB 2020. https://doi.org/10.1016/j.cels.2020.04.001
- Useful relevant videos
- Metastasis: How Cancer Spreads - from the NIH
- Clonal Evolution in Tumors and metastases - from Professor Ben Raphael at CGWI 2019
- Cancer Chromosomal Evolution in Metastases & Immune Escape - from Professor Charles Swanton CPM Annual Lecture 2018.
- Inferring Cellular Migrations in Metastatic Cancers - from Professor Ben Raphael at UCLA in 2018
- Phylogenetic Reconstruction of Clonal Evolution in Tumors and Metastases - from Professor Ben Raphael at B3D at Harvard University in 2018
- PEACE: Collecting cancer tissue to solve the riddle of metastasis - from Dr Mariam Jamal-Hanjani at ESMO 2018
- Machine Learning for Understanding Cancer Evolution - from Dr Roland Schwarz at MDC Berlin in 2019
 Close
Close