(Pre-July 2015)
Project Leaders: Dr Lee Butcher and Dr Dirk Paul
Background
Variation in DNA methylation has been associated with predisposition, progression and response to treatment of a broad range of clinical conditions such as cancer and autoimmune diseases (Feinberg A.P. (2007), Nature 447, 433-440). The aim of this project is to develop novel experimental and computational methods for the analysis of 5-methyl-cytosine (5-mC) and 5-hydoxymethyl-cytosine (5-hmC) using both genome-wide and targeted approaches.
Methylated DNA immunoprecipitation (MeDIP-Seq)
DNA methylation affects approximately 60-80% of the 28M CpG loci in the genome (Lister R. et al (2009), Nature 462, 315-322; Li Y. et al (2010) PLoS Biol. 8, e1000533). Given this vast, pliable landscape – and taking lessons from GWAS, in which genome-wide, hypothesis-free approaches often yielded surprising results – whole genome approaches to studying the methylation (co-called 'methylomes') are prudent. Here, local methylation events are evaluated against the background of methylome signals to separate signal from noise.
A number of techniques for generating whole-methylome datasets are available. The gold standard is Whole Genome Bisulfite Sequencing (WGBS), in which the methylated status of every cytosine in the genome can be resolved at single base pair resolution. Despite this benefit though, WGBS is time consuming, costly and (currently at least) inefficient, with 70-80% of sequencing reads providing little to no information (Ziller M. et al (2013) Nature 500, 477-481). One lower-cost alternative is Methylated DNA Immunoprecipitation (MeDIP), which involves co-incubation of DNA with an antibody raised against 5-mC along with magnetic beads, which form bead-antibody-methylated DNA complexes. These complexes are then separated from the unmethylated fraction and when coupled with next generation sequencing can assess more 50% of methylated CpG dinucleotides at 150- to 200-bp resolution, concomitant with sequence inset size.
This technique has been used to resolve the first DNA methylome (Down T.A. et al (2008), Nat. Biotechnol. 26, 779-785), map aberrant DNA methylation in malignant peripheral nerve sheath tumours (Feber A. et al (2011), Genome Res. 21, 515-524), and to locate differentially methylated regions associated with aging in mouse hematopoietic stem cells (Taiwo O. et al (2013), Epigenetics 8, 1114-1122).
Project Members:
- Lee Butcher
- Emanuele Libertini,
- Andrew Feber
Large-scale, targeted bisulphite sequencing using RainDrop BS-seq
Several techniques have been established to map DNA methylation in single CpG resolution at a selected subset of genomic regions of interest, for example to validate signals from epigenome-wide association studies (see: EWAS project page). Such methods include pyrosequencing, Sanger sequencing and high-resolution melting curve analysis. However, most of these have not been optimised for high-throughput applications. Emerging techniques that utilise next-generation DNA sequencing platforms are particularly promising for the large-scale, targeted bisulphite sequencing of genomic regions of interest.
RainDance Technologies developed a fully integrated enrichment system using microdroplet PCR that can also be coupled to next-generation sequencing platforms (Tewhey R. et al (2009), Nat. Biotechnol. 27, 1025-1031). The encapsulation of distinct PCR reactions in microdroplets enables the sensitive, specific and simultaneous amplification of up to 20,000 target loci using either unconverted or bisulphite-converted genomic DNA. We refined the approach into targeted RainDrop BS-seq and used it to validate a hypermethylation phenotype in isocitrate dehydrogenase (IDH) mutant chondrosarcoma (Guilhamon P. et al (2013), Nat. Commu. 4, 2166). Further, we recently presented a systematic assessment of RainDrop BS-seq as a method for large-scale, targeted bisulfite sequencing using a wide range of starting DNA quantity, quality and different cell types (Paul D.S. et al (2014), Epigenetics 9, 678-684).
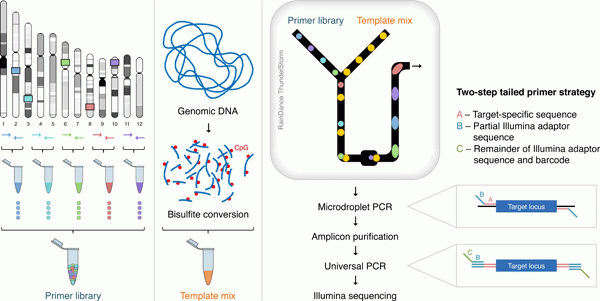
The custom primer panel for the genomic regions of choice is prepared by RainDance Technologies. The workflow comprises the following key steps: (1) bisulphite conversion of genomic template; (2) merger of picoliter-volume droplets of bisulphite-treated template with pre-made primer pair droplets (primer panel) on microfluidic chips; (3) pooled, thermal cycling of the PCR reactions (microdroplet PCR); (4) destabilization of droplets to release the PCR products; (5) purification of PCR products using magnetic beads; (6) incorporation of DNA sequencing barcodes through standard PCR (universal PCR), followed by purification of the PCR products and Illumina sequencing.
Project Members
- Dirk Paul
- Paul Guilhamon
- Pawan Dhami
- Stefan Stricker, Andrew Feber
 Close
Close




