Project Leader: Gareth Wilson
This project involved the development of computational tools for analysis of DNA methylation data from second generation sequencing platforms.
DNA Sequence Analysis
Our focus is on the analysis of MeDIP-seq, though we are also experienced in the analysis of RNA-seq, ChIP-seq, bis-seq and exome sequencing.
Originally, the Batman algorithm (Down et al., 2008) was used for our MeDIP analyses, including the first cancer methylome (Feber et al., 2011). Since then we have developed the MeDUSA pipeline (Wilson et al., 2012). The focus of which is to locate differentially methylated regions between cohorts.
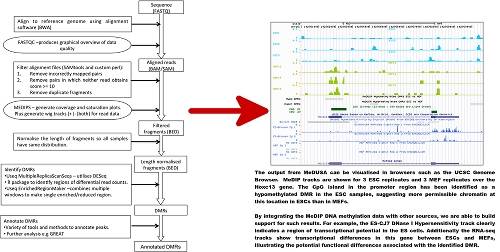
MeDUSA (Methylated DNA Utility for Sequence Analysis) brings together numerous software packages to perform a full analysis of MeDIP-seq data, including sequence alignment, quality control (QC), and determination and annotation of DMRs. MeDUSA utilises several applications from within the USeq software suite, and in turn uses the R Bioconductor package DESeq for differential count analysis. In addition, MeDUSA will control several other important functions from the alignment (BWA) and subsequent filtering (SAMtools), through generation of numerous quality control metrics (FastQC and MEDIPS), and finally preliminary annotation of the DMRs (utilising the capabilities of BEDTools). Download MeDUSA. Supporting data and associated files for the MeDUSA pipeline are also available from GigaScience .
A focus for future research within the group is on the integration of disparate datasets in order to elucidate a fuller understanding of the underlying biology and thus address fundamental questions associated with epigenetic regulation of mammalian cells.
References
1. Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al: A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 2008, 26:779-785.
2. Feber A, Wilson GA, Zhang L, Presneau N, Idowu B, Down TA, Rakyan VK, Noon LA, Lloyd AC, Stupka E, et al: Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res 2011, 21:515-524.
3. Wilson GA, Dhami P, Feber A, Cortazar D, Suzuki Y, Schulz R, Scchar P, Beck S: Resources for methylome analysis suitable for gene knockout studies of potential epigenome modifiers. Giga Science 2012, 1:3.
 Close
Close