New research leads the way in epigenomic studies
29 November 2016
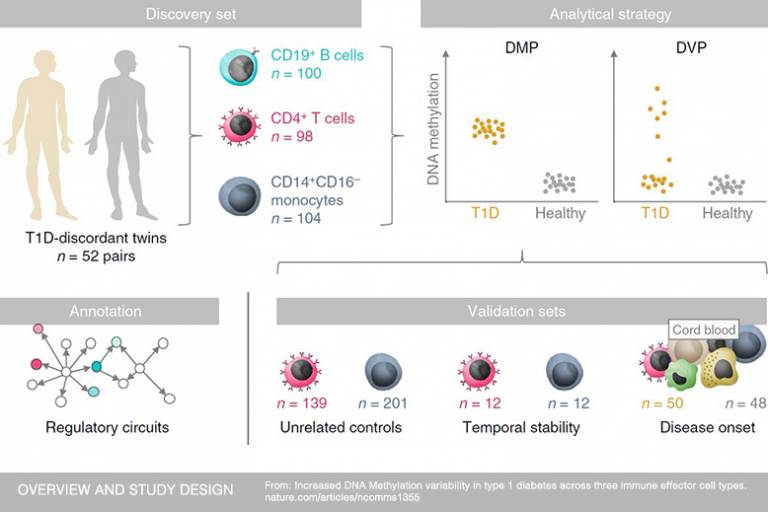
Research led by UCL and Queen Mary University of London (QMUL) scientists has identified epigenetic changes in three types of immune cell that could contribute to the development of type 1 diabetes. These changes were only found in patients with diabetes, opening the door to future biomarker development. The study, published today in Nature Communications, employed a unique experimental design that can now allow researchers to more confidently interpret results from epigenomic studies.
Type 1 diabetes is an autoimmune disease that results from the loss of insulin-producing cells in the pancreas. The dramatic increase in the incidence of the disease over recent years, particularly in children younger than five years of age, suggests that non-genetic factors have a major role to play. In this study, the researchers focused on epigenetic modifications – molecular changes induced by environmental cues that have no effect on the DNA sequence itself but affect how the DNA functions. For example, DNA methylation, the type of epigenetic modification studied here, can contribute to disease development and progression through its influence on gene expression.
The team performed an Epigenome-Wide Association Study (EWAS), designed to measure DNA methylation levels in large numbers of people to look at variations between those with and without a particular disease. They set out to investigate whether methylation patterns associated with type 1 diabetes in different cell types, with the aim of gaining a greater insight into the mechanisms driving the disease. However, meaningful interpretation of EWAS findings can be hampered by confounding factors, such as genetic differences between unrelated individuals and differences in blood samples (consisting of many different types of cell). Both of these factors can then render results inaccurate and so the team designed their study specifically to address these barriers.
“In this work, we aimed to determine differential DNA methylation in over 50 pairs of identical twins discordant for type 1 diabetes, meaning one twin has the disease and the other does not. Using identical twins is a clever way of eliminating genetic differences in order to gain a clearer picture of the specific role of epigenetic factors in the disease. In these twin pairs, we performed an EWAS in three types of immune cell known to act as key drivers in the disease process,” explains Dr Dirk Paul, former Research Associate in Professor Beck’s Medical Genomics Group at the UCL Cancer Institute, now at the University of Cambridge.
“We identified thousands of epigenetic marks across the three immune cell types that were only found in the twins with diabetes. These marks clustered near genes that play a role in immune cell metabolism and the cell cycle,” says Professor David Leslie from QMUL’s Blizard Institute and co-author of the study.
The scientists went one step further to determine the time point at which these epigenetic changes occurred. By investigating the DNA methylation patterns in cord blood of newborns who progressed to develop diabetes, the team provided evidence that the epigenetic changes likely emerged after birth.
The team hopes their experimental design, which was based on 772 genome-wide DNA methylation profiles, will now serve as a guide for future epigenetic studies.
This work is the result of an international collaboration as part of the International Human Epigenome Consortium (IHEC) of which the EU-FP7 Project BLUEPRINT is a member. This study was part-funded by the MRC.
Further information
- Research paper: ‘Increased DNA methylation variability in type 1 diabetes across three immune effector cell types’ Nature Communications
- Integrative Human Genomics Team (Dr Dirk Paul)
- Medical Genomics Group (Professor Stephan Beck)
- Professor David Leslie academic profile
- Professor Vardhman Rakyan academic profile
- Image: Figure 1. Overview and study design - graphic from ‘Increased DNA methylation variability in type 1 diabetes across three immune effector cell types’
Related news
BLUEPRINT celebrates major manuscript release
European epigenomes research project, BLUEPRINT, today announced the release of a collection of 26 publications in Cell, Cell Press-associated and other high-impact journals. The papers are part of a package of 41 publications by the International Human Epigenome Consortium (IHEC) of which BLUEPRINT is a member. Many key papers within the release are authored by Professor Stephan Beck and his scientific team from the Medical Genomics group at UCL Cancer Institute.
 Close
Close


