BLUEPRINT celebrates major manuscript release
17 November 2016

European epigenomes research project, BLUEPRINT, today announced the release of a collection of 26 publications in Cell, Cell Press-associated and other high-impact journals. The papers are part of a package of 41 publications by the International Human Epigenome Consortium (IHEC) of which BLUEPRINT is a member. Many key papers within the release are authored by Professor Stephan Beck and his scientific team from the Medical Genomics group at UCL Cancer Institute.
The EU-funded BLUEPRINT project is a large-scale research project consisting of 42 leading European universities, research institutes and industry entrepreneurs. Researchers in the project have focused on studying blood to produce a growing body of work on how the human genome can make blood cells, how those cells differentiate and what epigenetic changes occur to cause disease such as cancer and autoimmune disease.
Advancing epigenomic discoveries
One of the great mysteries in biology is how the many different cell types that make up our bodies are derived from a single cell and from one DNA sequence, or genome. We have learned a lot from studying the human genome, but have only partially unveiled the processes underlying cell determination. The identity of each cell type is largely defined by an instructive layer of molecular annotations on top of the genome – the epigenome – which acts as a blueprint unique to each cell type and developmental stage. Unlike the genome the epigenome changes as cells develop and in response to changes in the environment. Defects in the factors that read, write and erase the epigenetic blueprint are involved in many diseases. The comprehensive analysis of the epigenomes of healthy and abnormal cells will facilitate new ways to diagnose and treat various diseases, and ultimately lead to improved health outcomes.
These papers represent the most recent work of IHEC member projects from Canada, the European Union, Germany, Japan, Singapore, South Korea, and the United States. The collection of publications showcases the achievements and scientific progress made by IHEC in core areas of current epigenetic investigations.
“The collection of manuscripts impressively demonstrates how epigenetic information and analyses can help find answers to pressing questions related to the cellular mechanisms associated with complex human diseases,” says Professor Hendrik Stunnenberg, Radboud University, The Netherlands and former Chair of the IHEC International Scientific Steering Committee and coordinator of the BLUEPRINT project.
“BLUEPRINT has produced more new science and more understanding of blood cell disease than we could have imagined at the outset. We have made the data of >1000 datasets publicly available, moreover, we have forged an alliance of researchers and innovative companies from around Europe and, working closely with international partners, we already see results that, in time, will improve the lives of patients.”
“BLUEPRINT has been a fantastically collaborative project to show case many aspects of the innovative epigenomic research conducted at UCL, from technology development to insights into biology and disease” says Professor Stephan Beck, Professor of Medical Genomics, whose research group led or contributed to nine of the publications.
UCL contributions to IHEC package
- 1. Saturation analysis for whole-genome bisulfite sequencing data
-
Libertini E, Heath SC, Hamoudi RA, Gut M, Ziller MJ, Herrero J, Czyz A, Ruotti V, Stunnenberg HG, Frontini M, Ouwehand WH, Meissner A, Gut IG, Beck S. Saturation analysis for whole-genome bisulfite sequencing data. Nature Biotechnology 2016, http://www.nature.com/nbt/journal/v34/n7/full/nbt.3524.html
- 2. Information recovery from low coverage whole-genome bisulfite sequencing
-
Libertini E, Heath SC, Hamoudi RA, Gut M, Ziller MJ, Czyz A, Ruotti V, Stunnenberg HG, Frontini M, Ouwehand WH, Meissner A, Gut IG, Beck S. Information recovery from low coverage whole-genome bisulfite sequencing. Nature Communications 2016, http://www.nature.com/ncomms/2016/160627/ncomms11306/full/ncomms11306.html
- 3. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications
-
Bock C, Halbritter F, Carmona FJ, Tierling S, Datlinger P, Assenov Y, Berdasco M, Bergmann AK, Booher K, Busato F, Campan M, Dahl C, Dahmcke CM, Diep D, Fernández AF, Gerhauser C, Haake A, Heilmann K, Holcomb T, Hussmann D, Ito M, Kläver R, Kreutz M, Kulis M, Lopez V, Nair SS, Paul DS, Plongthongkum N, Qu W, Queirós AC, Reinicke F, Sauter G, Schlomm T, Statham A, Stirzaker C, Strogantsev R, Urdinguio RG, Walter K, Weichenhan D, Weisenberger DJ, Beck S, Clark SJ, Esteller M, Ferguson-Smith AC, Fraga MF, Guldberg P, Hansen LL, Laird PW, Martín-Subero JI, Nygren AO, Peist R, Plass C, Shames DS, Siebert R, Sun X, Tost J, Walter J, Zhang K. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nature Biotechnology 2016, http://www.nature.com/nbt/journal/v34/n7/full/nbt.3605.html
- 4. Increased DNA methylation variability in type 1 diabetes across three immune effector cell types
-
Dirk S. Paul, Andrew E. Teschendorff, Mary A. N. Dang, Robert Lowe, Mohammed I. Hawa, Simone Ecker, Stephanie Cunningham, Alexandra R. Fouts, Anita Ramelius, Frances Burden, Samantha Farrow, Sophia Rowlston, Karola Rehnstrom, Mattia Frontini, Kate Downes, Stephan Busche, Warren Cheung, Bing Ge, Marie-Michelle Simon, David Bujold, Tony Kwan, Guillaume Bourque, Avik Datta, Ernesto Lowy, Laura Clarke, Paul Flicek, Emanuele Libertini, Simon Heath, Marta Gut, Ivo G. Gut, Willem H. Ouwehand, Tomi Pastinen, Nicole Soranzo, Sabine E. Hofer, Beate Karges, Thomas Meissner, Bernhard O. Boehm, Corrado Cilio, Helena Elding Larsson, Åke Lernmark, Andrea K. Steck, Vardhman K. Rakyan, Stephan Beck & R. David Leslie. Increased DNA methylation variability in type 1 diabetes across three immune effector cell types. Nature Communications 2016, http://dx.doi.org/10.1038/ncomms13555
- 5. A human variation panel of genetic influences on epigenomes and transcriptomes in three immune cells
-
Lu Chen, Bing Ge, Francesco Paolo Casale, Louella Vasquez, Tony Kwan, Diego Garrido, Stephen Watt, Ying Yang, Simone Ecker, Avik Datta, David Richardson, Frances Burden, Daniel Mead, Alice L. Mann, Jose Maria Fernandez, Kousik Kundu, Sophia Rowlston, Steven P. Wilder, Samantha Farrow, Xiaojian Shao, John J. Lambourne, Adriana Redensek, Cornelis A. Albers, Vyacheslav Amstislavskiy, Sofie Ashford, Kim Berentsen, Lorenzo Bomba, Guillaume Bourque, David Bujold, Stephan Busche, Maxime Caron, Shu-Huang Chen, Warren Cheung, Oliver Delaneau, Emmanouil . Dermitzakis, Heather Elding, Irina Colgiu, Frederik O. Bagger, Paul Flicek, Ehsan Habibi, Valentina Iotchkova, Eva Janssen-Megens, Bowon Kim, Hans Lehrach, Ernesto Lowy, Amit Mandoli, Filomena Matarese, Matthew T. Maurano, John A. Morris, Vera Pancaldi, Farzin Pourfarzad, Karola Rehnstrom, Augusto Rendon, Thomas Risch, Nilofar Sharifi, Marie-Michelle Simon, Marc Sultan, Alfonso Valencia, Klaudia Walter, Shuang-Yin Wang, Mattia Frontini, Stylianos E. Antonarakis, Laura Clarke, Marie-Laure Yaspo, Stephan Beck, Roderic Guigo, Daniel Rico, Joost H. A. Martens, Willem H. Ouwehand, Taco W. Kuijpers, Dirk S. Paul, Hendrick G. Stunnenberg, Oliver Stegle, Kate Downes, Tomi Pastinen, Nicole Soranzo. A human variation panel of genetic influences on epigenomes and transcriptomes in three immune cells. Cell 2016, http://dx.doi.org/10.1016/j.cell.2016.10.026
- 6. The allelic landscape of human blood cell trait variation
-
William J. Astle, Heather Elding, Tao Jiang, Dave Allen, Dace Ruklisa, Alice L. Mann, Daniel Mead, Heleen Bouman, Fernando Riveros-Mckay, Myrto A. Kostadima, John J. Lambourne, Suthesh Sivapalaratnam, Kate Downes, Kousik Kundu, Lorenzo Bomba, Kim Berentsen, John R. Bradley, Louise C. Daugherty, Olivier Delaneau, Stephen F. Garner, Luigi Grassi, Matthias Haimel, Eva M. Janssen-Megens, Anita Kaan, Mihir Kamat, Bowon Kim, Amit Mandoli, Jonathan Marchini, Joost H.A. Martens, Stuart Meacham, Karyn Megy, Jared O’Connell, Romina Petersen, Nilofar Sharifi, Simon M. Sheard, James R. Staley, Salih Tuna, Martijn van der Ent, Shuang-Yin Wang, Eleanor Wheeler, Steven P. Wilder, Valentina Iotchkova, Carmel Moore, Jennifer Sambrook, Hendrik G. Stunnenberg, Emanuele Di Angelantonio, Stephen Kaptoge, Taco W. Kuijpers, Enrique Carrillo-de-Santa- Pau, David Juan, Daniel Rico, Alfonso Valencia, Lu Chen, Bing Ge, Louella Vasquez, Tony Kwan, Diego Garrido-Martín, Stephen Watt, Ying Yang, Roderic Guigo, Stephan Beck, Dirk S. Paul, Tomi Pastinen, David Bujold, Guillaume Bourque, Mattia Frontini, John Danesh, David J. Roberts, Willem H. Ouwehand, Adam S. Butterworth, Nicole Soranzo. The allelic landscape of human blood cell trait variation. Cell 2016, http://dx.doi.org/10.1016/j.cell.2016.10.042
- 7. eFORGE: a tool for identifying cell type-specific signal in epigenomic data
-
Charles E. Breeze, Dirk S. Paul, Jenny van Donge, Lee M. Butcher, John C. Ambrose, James E. Barrett, Robert Lowe, Vardhman K. Rakyan, Valentina Iotchkova, Mattia Frontini, Kate Downes, Willem H. Ouwehand, Jonathan Laperle, Pierre-Étienne Jacques, Guillaume Bourque, Anke K. Bergmann, Reiner Siebert, Edo Vellenga, Sadia Saeed, Filomena Matarese, Joost H.A. Martens, Hendrik G. Stunnenberg, Andrew E. Teschendorff, Javier Herrero, Ewan Birney, Ian Dunham, Stephan Beck. eFORGE: a tool for identifying cell type-specific signal in epigenomic data. Cell Reports 2016, http://www.cell.com/cell-reports/fulltext/S2211-1247(16)31479-6
- 8. From profiles to function in epigenomics
-
Stefan H. Stricker, Anna Köferle, Stephan Beck. From profiles to function in epigenomics. Nature Reviews Genetics 2016, http://www.dx.doi.org/10.1038/nrg.2016.138
- 9. The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery
-
BLUEPRINT Consortium: The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery. Cell 2016
Further information
- BLUEPRINT
- Cell: Insights from the International Human Genome Consortium
- IHEC
- Medical Genomics Group (Professor Stephan Beck)
- BLUEPRINT videos
Related news
Powerful analysis tool for epigenomic studies developed
Research published in Cell Reports highlights a powerful new data analysis tool that could vastly improve the interpretation of data gathered from epigenomic studies. The scientific team, led by UCL, have developed the eFORGE tool, which offers a new way to identify cell types that underlie complex human disease.
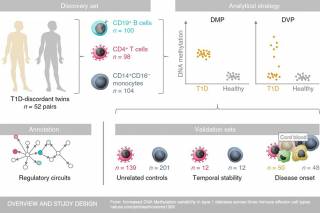
New research leads the way in epigenomic studies
Research led by UCL and Queen Mary University of London (QMUL) scientists has identified epigenetic changes in three types of immune cell that could contribute to the development of type 1 diabetes.
 Close
Close


