Genome sequencing reveals complexity of undifferentiated sarcoma
18 March 2019
A team of researchers has unpicked the complexity of an unusual group of rare tumours, a subset of which might be candidates for immune checkpoint inhibitor therapies.
The study, published in the journal Cancer Cell, provides insight into the biology and evolution of undifferentiated sarcoma and paves the way for possible treatment options and further research into the disease.
Undifferentiated sarcomas are a heterogeneous group of soft-tissue tumours. The label is generally applied when other diagnoses have been excluded, acting as a ‘wastepaper basket’ for sarcomas which have defied classification. They are often detected late, and prognosis is generally poor – median survival for those with metastatic disease is about 12 months.
Because of their rarity and diverse nature, undifferentiated sarcomas have been little studied. However, classical chromosomal studies suggest that they are highly complex.
To gain a clearer picture, a team led by Dr Nischalan Pillay and colleagues from UCL, the Wellcome Sanger Institute and the Francis Crick Institute analysed a unique series of undifferentiated sarcoma samples. These tumour samples, generously donated by patients for research, were collected over a decade by Professor Adrienne Flanagan at the Royal National Orthopaedic Hospital as part of the London Sarcoma Service.
"We used a range of methods to analyse 76 tumours, of which whole genome sequencing was by far the most informative. The sequence data revealed that tumours could be broken down into multiple groups based on the nature of their genetic abnormalities," said Dr Pillay.
Notably, in around 15% of the tumours analysed, simple point mutations were the most commonly observed genetic changes. Several of these changes were in genes affecting the immune response to tumours, and the prognosis of patients with this type of tumour was markedly better than that of other patients.
"These findings suggest that patients found to carry this type of mutation pattern might benefit from anticancer drugs targeting the immune system, such as checkpoint inhibitors. With NHS patients having the option of having their cancers whole genome sequenced in the coming months, you can see why these findings are valuable - we would be able to identify patients who would be most suitable for clinical trials of these drugs," said Dr Pillay.
The team developed an innovative method to characterise the tumours’ genomic abnormalities and were able to determine the order in which the changes occurred. These analyses confirmed that undifferentiated sarcomas use duplications of their genomes to evolve, and identified a range of pathways by which cells acquired their cancerous properties. Another common feature was a dramatic shattering of chromosomes with redistribution of chunks of DNA to other parts of the genome which altered genes that cause cancer.
Using this method they were able to show that, strikingly, the evolution of tumours took place over wildly different timeframes. In some cases, genome duplication took place decades before cancers were detected, while in other patients, duplications or rearrangements very rapidly led to unrestrained cancer growth.
“The complexity of undifferentiated sarcoma genomes necessitated the application of state-of-the-art and novel computational methods to reveal the complex biology underlying these tumours. The insights gained shed important light on potential future treatment and research avenues for this rare and under studied tumour type,” said first author Dr Christopher Steele (UCL Cancer Institute).
Co-author Dr Peter Van Loo (The Francis Crick Institute) said: "This study paints a comprehensive picture of the genomic landscape of undifferentiated sarcomas, showing which are the key genes and complex mutational processes underlying the disease."
This study demonstrates how comprehensive multi-omic molecular analysis of a well defined tumour group when interrogated alongside clinical outcome provides better prognostic indicators for patients, and will serve as a model for future studies. "
Professor Adrienne Flanagan, Professor of Musculoskeletal Pathology (UCL) and Consultant Histopathologist (RNOH)“
This work was supported by Cancer Research UK, the UK Medical Research Council and the Wellcome Trust.
Further information
- Research paper: Undifferentiated sarcomas develop through distinct evolutionary pathways. Cancer Cell.
- Pillay Lab - Sarcoma Biology and Genomics Research Group
- Van Loo Lab - Cancer Genomics
- Professor Adrienne Flanagan academic profile
- Royal National Orthopaedic Hospital (RNOH)
Image
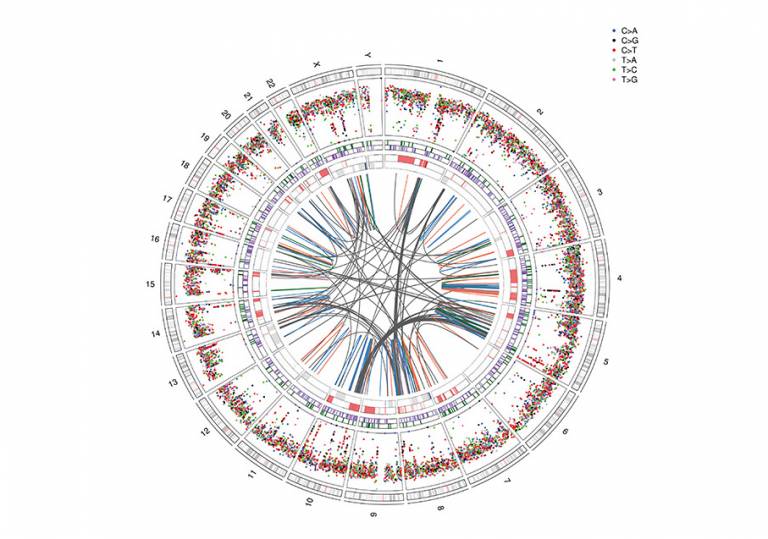
- Ideogram of the genomic complexity seen in USARC.
- Image detail
An integrated examination of undifferentiated sarcoma(USARC) revealed evolutionary trajectories defined by altered copy number states. The coordinates of chromosomes are shown in the outer ring. The second, third and fourth outermost rings represent mutations and alterations of DNA copy number as a function of genomic coordinates. The centre arcs highlight the complex rearrangements between chromosomes. Immune "hot" molecular subtypes were identified which could open up avenues for immune checkpoint blockade.
Media contact
Henry Kilworth +44 207 679 5296
h.killworth@ucl.ac.uk
 Close
Close

