Unravelling the dynamics of small-molecule interactions with intrinsically disordered proteins
1 February 2024
A new study from UCL entitled "Picosecond Dynamics of a Small Molecule in Its Bound State with an Intrinsically Disordered Protein," has been recently published in the Journal of the American Chemical Society.
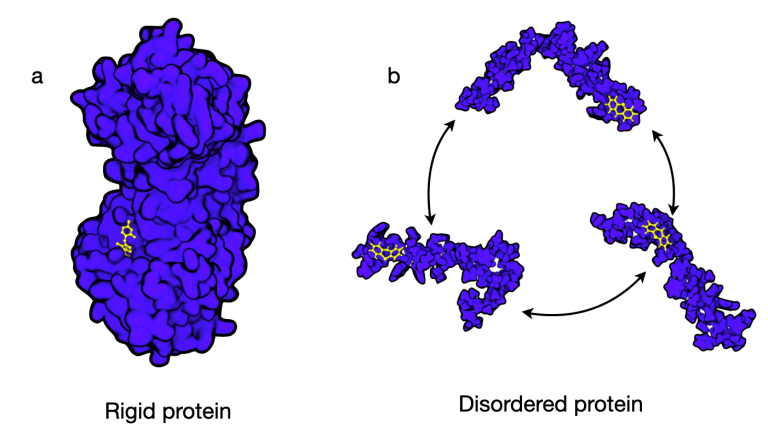
Many processes in biology depend on how large biomolecules, called proteins, interact with smaller chemicals or 'small molecules'. These interactions are often described in terms of a ‘lock-and-key’ binding, whereby small molecules (keys) fit snugly into ridges or groves on the protein surface (locks). While about 2/3 of proteins adopt well-defined 3D shapes associated with a specific function, many proteins, including those involved in human-, viral-, and plant biology, never adopt a single shape and instead rapidly interconvert between many shapes.
These proteins, called 'disordered proteins', lack long-lived ‘locks’ to which small molecule ‘keys’ can bind, making them difficult to target in drug discovery programmes using traditional methods. In fact, disordered proteins have been considered "undruggable" due to their lack of classical long-lived binding pockets for drugs.
Frequently, scientists will use a method called nuclear magnetic resonance (NMR) spectroscopy to study small-molecule interactions with proteins. Traditionally, however, these methods have been insensitive to binding between small molecules and disordered proteins. BBSRC Discovery Fellow Gabriella Heller and Professor D. Flemming Hansen in the Department of Structural and Molecular Biology (Division of Biosciences) have recently developed a new NMR-based strategy based on the effective linewidth of 19F signals to detect the binding between a small molecule and a disordered protein. Using this approach, they discovered that a small molecule not only interacts with a disordered protein but does so in exceptionally dynamic ways, in stark contrast to the traditional ‘lock-and-key’ binding mechanism.
Dr Heller, the first author on the manuscript states ‘Not only can we experimentally detect binding to these “undruggable” proteins, but we can also learn something interesting about the mechanism of this interaction - the small molecule moves very quickly in the bound state - it’s quite unusual’.
Professor D. Flemming Hansen, the lead researcher, expresses excitement because ‘this study opens a new interdisciplinary scientific discipline, which links physics, chemistry, biology, as well as medicine, and where focus the next years will be on understanding and exploiting the mechanisms by which small molecules interact to disordered proteins.’
Further Information:
- Dr Gabriella Heller staff profile
- Professor Flemming Hansen staff profile
- Publication: Picosecond Dynamics of a Small Molecule in Its Bound State with an Intrinsically Disordered Protein
- Research Group website
- Research Department of Structural and Molecular Biology
- Division of Biosciences
Image:
Targeting disordered and flexible proteins represents a novel drug discovery strategy. (a) Conformations of a rigid protein with well-defined grooves and pockets, which can act as binding sites. (b) A disordered protein, which rapidly interconverts between states and generally considered not to have ‘traditional’ drug binding sites.
 Close
Close

