Functional profiling of long intergenic non-coding RNAs in fission yeast
27 April 2022
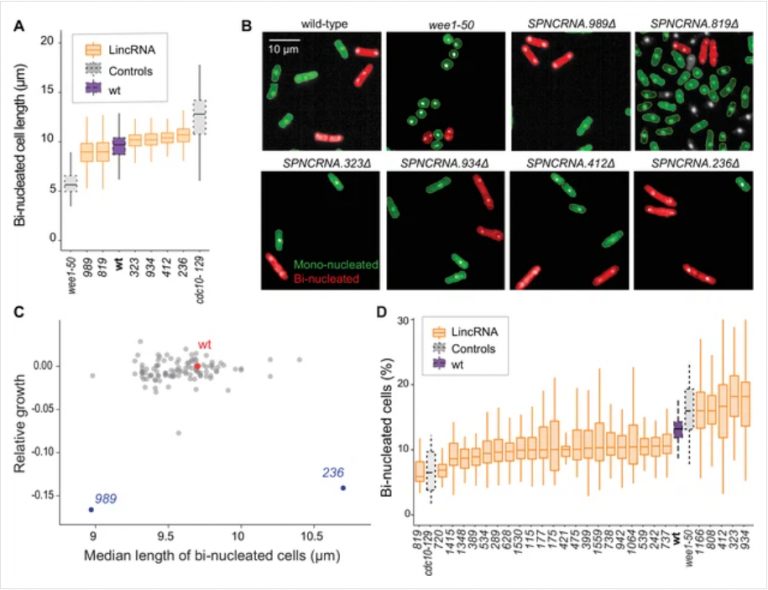
Using seamless CRISPR/Cas9-based genome editing, we deleted 141 lincRNA genes to broadly phenotype these mutants, together with 238 diverse coding-gene mutants for functional context. We applied high-throughput colony-based assays to determine mutant growth and viability in benign conditions and in response to 145 different nutrient, drug, and stress conditions.
These analyses uncovered phenotypes for 47.5% of the lincRNAs and 96% of the protein-coding genes.
These rich phenomics datasets associate lincRNA mutants with hundreds of phenotypes, indicating that most of the lincRNAs analysed exert cellular functions in specific environmental or physiological contexts. This study provides the groundwork to further dissect the roles of these lincRNAs in the relevant conditions.
Functional profiling of long intergenic non-coding RNAs in fission yeast | eLife (elifesciences.org)
 Close
Close

