Huge bacteria-killing viruses blur the boundaries defining life
13 February 2020
Hundreds of unusually large, bacteria-killing viruses with capabilities normally associated with living organisms, have been identified by a team involving UCL, blurring the line between viruses and living microbes.
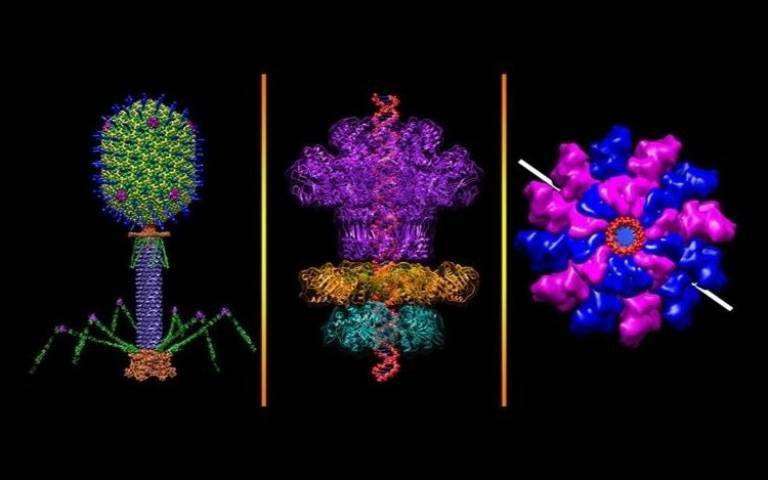
These phages — short for bacteriophage, so-called because they “eat” bacteria — are of a size and complexity considered typical of life, carry numerous genes normally found in bacteria, and use these genes against their bacterial hosts and other viruses, as reported in Nature.
“In the DNA of these phages, we found parts of the system that bacteria use to fight viruses. It appears that some of these phages are using this system to target other viruses,” said co-author Professor Joanne Santini (UCL Structural & Molecular Biology).
“There are likely phage wars – viruses targeting other viruses – going on inside the gut microbiome of humans.”
The researchers, led by University of California, Berkeley, found these huge phages by scouring a large database of DNA that they generated from nearly 30 different Earth environments, ranging from the guts of premature infants and pregnant women to a Tibetan hot spring, a South African bioreactor, hospital rooms, oceans, lakes and deep underground.
Professor Santini of UCL led analysis of phages from people in Bangladesh who had been drinking arsenic-tainted water, and phages from an arsenic-contaminated mine in northern Canada.
Altogether the researchers identified 351 different huge phages, all with genomes four or more times larger than the average genome of viruses that prey on bacteria.
Among these is the largest bacteriophage discovered to date: its genome, 735,000 base-pairs long, is nearly 15 times larger than the average phage and is much larger than genomes of many bacteria.
Lead author Professor Jill Banfield (UC Berkeley) said: “We are exploring Earth's microbiomes and sometimes unexpected things turn up. These viruses of bacteria are a part of biology, of replicating entities, that we know very little about.”
“These huge phages bridge the gap between non-living bacteriophage, on the one hand, and bacteria and archaea. There definitely seems to be successful strategies of existence that are hybrids between what we think of as traditional viruses and traditional living organisms.”
Within the DNA of some huge phages are parts of the CRISPR system that bacteria use to target viruses. It’s likely that once these phages inject their DNA into bacteria, the viral CRISPR system augments the CRISPR system of the host bacteria, probably mostly to target other viruses. Some of them also have CRISPR arrays, which are areas of the bacterial genome where snippets of viral DNA are stored for future reference, allowing bacteria to recognise returning phage and mobilise their proteins to target and cut them up.
UC Berkeley graduate student Basem Al-Shayeb, co-first author of the paper, said: “It is fascinating how these phages have repurposed this system we thought of as bacterial or archaeal to use for their own benefit against their competition, to fuel warfare between these viruses.”
One of the huge phages, analysed by Professor Santini from faecal samples in Bangladesh, is able to make a protein analogous to the Cas9 protein that is part of the revolutionary tool CRISPR-Cas9 that researchers have adapted for gene-editing in recent years.
The new findings could have implications for human disease, as phages may carry damaging genes, such as those coding for antimicrobial resistance, into the bacteria that colonise humans.
In a previous Nature paper last year, Professors Banfield and Santini reported that some of the largest phages, a group called Lak phages, can be found in people’s guts.*
Professor Santini said: “We were surprised to find that the Lak phages, found in the guts of people, bore more resemblance to phages found in pigs than in baboons, which suggests they were likely transmitted from pigs to people.”
She added: “Huge phages are prevalent across the globe, in people, animals and other environments. We still have a lot to learn about these under-studied viruses – for one, to learn about how they may help or hinder human health.”
While most of the genes in these huge phages code for unknown proteins, the researchers were able to identify genes that code for proteins called ribosomes, which are typically found in bacteria or archaea but not in viruses.
Co-first author Rohan Sachdeva (UC Berkeley) said: “Typically what separates life from non-life is to have ribosomes and the ability to do translation; that is one of the major defining features that separate viruses and bacteria, non-life and life. Some large phages have a lot of this translational machinery, so they are blurring the line a bit.”
Lead funding for the study came from the US National Institutes of Health.
Links
- Research paper in Nature
- Professor Joanne Santini’s academic profile
- UCL Structural & Molecular Biology
Image
- 'Bacteriophage' (not from study). Credit: Victor Padilla-Sanchez, The Catholic University of America, Source: National Institutes of Health on Flickr
Media contact
Chris Lane
Tel: +44 (0)207 679 9222
Email: chris.lane [at] ucl.ac.uk
 Close
Close

