Jan Bieschke
Research Synopsis
We aim to understand the central problem in the prion mechanism; what is the change in shape that distinguishes normal prion protein, PrPC , from its rogue form, PrPSc , and how does it come about? Specifically, we are asking what the structural causes are for becoming a prion, what the common drivers are for their replication, and whether the same principles underlie other diseases, both inside the central nervous system and in the rest of the body.
We combine biophysical techniques with nanoscopic imaging to study the structure, folding and dynamics of prions, both in isolation and with likely binding partners, in order to develop new strategies against prion diseases. We have developed nanoscopic imaging to observe amyloid structures over extended times through transient amyloid binding (TAB) of dye molecules.
A population of prions can have individual prion particles with different folds, which template different fibril structures during elongation. These individual prion particles form a ‘quasi-species’, where individual particles compete for substrate and where the replication environment shapes the replication ‘fitness’ of each prion particle. Fibril structure can ‘mutate’ in mid-growth as shown in the movie.
Prions therefore recapitulate the basic molecular principles of Darwinian evolution in a system, which does not contain genetic information coded by DNA or RNA. (Sun et al. ACS Nano 2023)
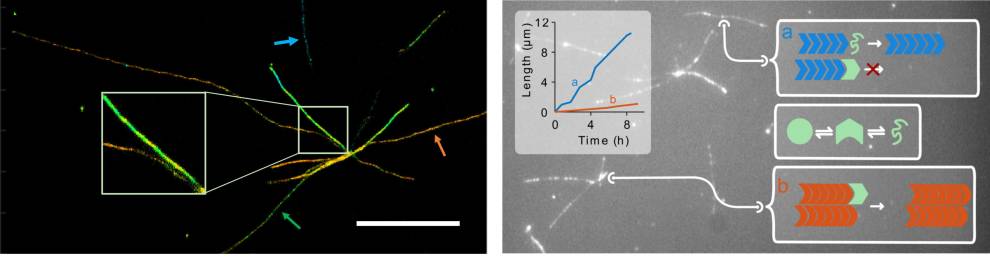
Growth of a single PrP fibril, which changes conformation in mid growth causing a switch from straight to helical morphology.
Direct Observation of Competing Prion Protein Fibril Populations with Distinct Structures and Kinetics
Y Sun, K Jack, T Ercolani, D Sangar, L Hosszu, J Collinge, J Bieschke ACS nano 2023
Loss of residues 119–136, including the first β-strand of human prion protein, generates an aggregation-competent partially “open” form
LLP Hosszu, D Sangar, M Batchelor, E Risse, AM Hounslow, J Collinge, J Bieschke. Journal of Molecular Biology 2023
Brazilin removes toxic alpha-Synuclein and seeding competent assemblies from Parkinson brain by altering conformational equilibrium
GR Nahass, Y Sun, Y Xu, M Batchelor, M Reilly, I Benilova, N Kedia, ... ,J Bieschke. Journal of molecular biology 2021
Structural effects of the highly protective V127 polymorphism on human prion protein
LLP Hosszu, R Conners, D Sangar, M Batchelor, EB Sawyer, S Fisher, ...Communications Biology 2020
 Close
Close


