Statistical modelling could be used to refine imaging genetics
14 March 2018
A paper published by UCL researchers in PNAS outlines an experimental imaging-genetics approach for investigating the genetic underpinnings of brain atrophy in Alzheimer's disease.
There are around 20,000 genes in the human genome. Understanding these genes and how they might indicate predisposition to certain diseases is an increasing research focus.
In neuroimaging specifically there is strong evidence to suggest that genes may hold the key to understanding neurodegenerative and neurological conditions for which we currently can't predict or treat effectively. However, due to the sheer volume of scale in the human genome current studies normally focus on predefined regions of the brain or genes of interest.
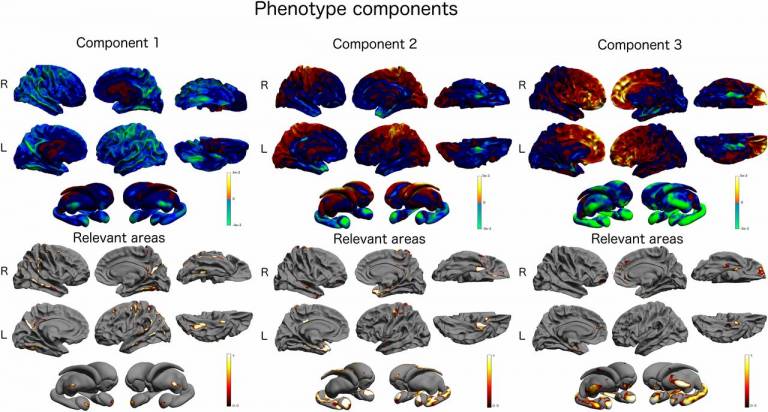
Researchers from the Translational Imaging Group, UCL Institute of Neurology and the Imaging Genetics Center at the University of Southern California have published results on a novel statistical approach to imaging-genetics which provides an efficient way of examining the whole brain versus the whole genome in order find key disease indicators which may have been previously overlooked. This method aims to remove bias by offering a feasible, quick method of looking at the information comprehensively.
To illustrate their method the research team took a dataset of 639 individuals (either Alzheimer's patients or healthy controls) from the Alzheimer's Disease Neuroimaging Initiative (ADNI) to analyse genetic variation.
The first stage of the process conducted an integrated analysis of cortical thickness of the brain measured at about 350,000 points and over 1 million genetic markers to hone in on just about 120 genetic regions.
The underlying statistical method, known as partial least squares, aims to find combinations of genetic markers that are maximally correlated to combinations of cortical thickness measures across the brain. Functional prioritisation, a process which analyses the genetic regions and the associated genes for any known relevant molecular biologic functional using public databases, was then applied to refine the number to just 14 candidate genes.
These genes of interest were then validated against another dataset from the ADNI containing 553 individuals. Within the second data set it was known which individuals displaying signs of mild cognitive impairment (MCI) would go on to develop Alzheimer's, providing a reliable benchmark to test their analysis against.
The results found that the gene TRIB3 (tribbles pseudokinase 3) was linked to the stereotypical pattern of grey matter loss, a key indicator linked to Alzheimer's.
It is hoped the methodology would provide enough statistical evidence to demonstrate the role this gene plays to be of interest in future studies into Alzheimer's.
The method provides a resource efficient way to unlock large genome databases without the need for high powered supercomputers. The work could also act as a roadmap for additional statistical analysis in the imaging-genetics field, streamlining the areas of focus for a number of diseases in this rapidly growing field of study.
Dr Andre Altmann comments "With this method we were able to analyse the "whole brain" and "whole genome" data without any prior assumptions on our notebooks. Furthermore, we are currently extending the model to analyse data that is not stored in one place but is distributed around the world. It will be exciting to see results on other datasets comprising tens of thousands of subjects."
Links
- Full paper, "Susceptibility of brain atrophy to TRIB3 in Alzheimer's disease, evidence from function prioritization in imaging genetics," PNAS
- UCL Institute of Neurology
- Translational Imaging Group
- Imaging Genetics Center of the University of Southern California
- Alzheimer's Disease Neuroimaging Initiative
- Experimental material is available on figshare.
 Close
Close

