New publication in Journal of Biological Chemistry for Ketteler Lab
In all eukaryotic cells, ubiquitin is a major post-translational modification that tags proteins for degradation. In Journal of Biological Chemistry, the Ketteler Lab describes a novel modification that involves proteins that are structurally similar to ubiquitin, but distinct in terms of their sequence. These proteins are involved in a distinct degradation system - autophagy. The researchers discover that proteins in cells are tagged with this ubiquitin-like modification in a very similar manner to ubiquitin-conjugation. The functional relevance is not clear at the moment, but implies that other ubiquitin-related processes exist in cells, thus expanding the complexity of post-translational modifications with this family of proteins.
Abstract from the article:
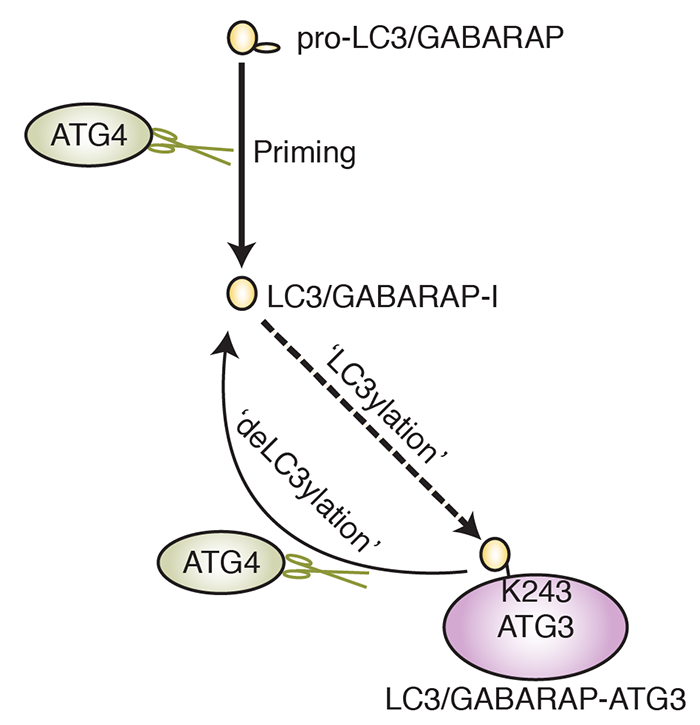
Microtubule-associated protein 1 light chain 3 alpha (LC3)/GABA type A receptor–associated protein (GABARAP) comprises a family of ubiquitin-like proteins involved in (macro)autophagy, an important intracellular degradation pathway that delivers cytoplasmic material to lysosomes via double-membrane vesicles called autophagosomes. The only currently known cellular molecules covalently modified by LC3/GABARAP are membrane phospholipids such as phosphatidylethanolamine in the autophagosome membrane. Autophagy-related 4A cysteine peptidase (ATG4) proteases process inactive pro-LC3/GABARAP before lipidation, and the same proteases can also deconjugate LC3/GABARAP from lipids. To determine whether LC3/GABARAP has other molecular targets, here we generated a preprocessed LC3B mutant (Q116P) that is resistant to ATG4-mediated deconjugation. Upon expression in human cells and when assessed by immunoblotting under reducing and denaturing conditions, deconjugation-resistant LC3B accumulated in multiple forms and at much higher molecular weights than free LC3B. We observed a similar accumulation when preprocessed versions of all mammalian LC3/GABARAP isoforms were expressed in ATG4-deficient cell lines, suggesting that LC3/GABARAP can attach also to other larger molecules. We identified ATG3, the E2-like enzyme involved in LC3/GABARAP lipidation, as one target of conjugation with multiple copies of LC3/GABARAP. We show that LC3B–ATG3 conjugates are distinct from the LC3B–ATG3 thioester intermediate formed before lipidation, and we biochemically demonstrate that ATG4B can cleave LC3B–ATG3 conjugates. Finally, we determined ATG3 residue K243 as an LC3B modification site. Overall, we provide the first cellular evidence that mammalian LC3/GABARAP post-translationally modifies proteins akin to ubiquitination (‘LC3ylation’), with ATG4 proteases acting like deubiquitinating enzymes to counteract this modification (‘deLC3ylation’).
 Close
Close