Group Leader: Professor Tariq Enver
Research
Because of their unique biological properties and potential medical importance, stem cells have attracted major scientific, commercial and public interest (Weissman et al, Annu Rev Cell Dev Biol; 17:387-403, 2001). The cell fate options that confront stem cells include self-renewal, differentiation and lineage-specification, programmed cell death and quiescence. Within haematopoiesis, constitutive dysregulation of the balance between these processes is an essential feature of leukaemogenesis. A key challenge is to understand how the different cell fates that confront stem and progenitor cells are selected and co-ordinated; adoption of a given fate must be coupled to appropriate suppression of alternative fates. While much still remains to be understood about the nature of the molecular pathways involved in the regulation of stem and progenitor cell fate, it is generally accepted that transcription factors are key intrinsic regulators of these decisions, through modulation of overlapping or linked networks of transcription factors (Enver and Greaves, Cell; 94:9-12, 1998, Shivdasani and Orkin, Blood; 87:4025-4039, 1996, Rothenberg et al, Dev Biol; 246:29-44, 2002). Figure 1. We have used multiple genome-wide and gene-specific approaches to examine the molecular mechanisms that regulate gene expression and consequently cell fate in haemopoietic mulipotential progenitor cells and their more differentiated progeny. In parallel, we have explored how chimaeric transcription factors associated with leukaemia impact the transcriptional circuitry and behaviour of haemopoietic stem and progenitor cells.
Our studies of normal cell fate control have been underpinned by the generation of a data-resource containing gene expression and microRNA profiles, TF and histone ChIP data as well as perturbation data and functional readouts derived from murine multipotential progenitors undergoing self-renewal and differentiation. The resource is complemented by related datasets from mouse and human primary and leukaemic cells as well as embryonic stem cells. At a simple level this resource has underpinned gene discovery studies of novel regulators of stem and progenitor cell fate such as Nov/CCN3; an essential regulator of human cord blood derived stem cells (Gupta et al 2007), Cited -2; a stem regulator shared with ES cells (Kranc et al 2009) and MLLT3, a novel player in the erythroid lineage (Pina et al 2008). A greater challenge has been to assemble these and related regulators into transcriptional networks and model their behaviour (Enver et al 2009, Graf and Enver 2010). In this respect we have developed novel models of GATA-PU.1 interaction (Huang et al 2007, Chickarmane et al 2009) and are currently developing a stochastic model based on single cell molecular and behavioural analysis that affords inference of the rules of lineage commitment in the blood system. The concepts and approaches developed have applicability in other stem cell systems, most notably ESC, and have informed our attempts to develop systems level approaches to leukaemia (see below).
Our current work is cancer biology based on the premise that understanding the pathogenesis of malignancy requires knowledge of (i) the nature of the underlying mutations, (ii) the cellular compartments in which they arise and (iii) their functional impact and its dependence on cellular context. We are exploring these issues in the context of t(12;21) associated childhood ALL. In this paradigmatic disorder, the fusion oncogene TEL-AML1 that encodes a chimaeric transcription factor constitutes an initiating or ‘first hit’ mutation. The identities of common co-operating genes in this disease (eg TEL1 and Pax5) are known, and significant insights into the cellular hierarchies of the pre-leukaemic and leukaemic phases have been made. Using a combination of experimental modelling and detailed analysis of patient derived samples, we are examining the nature and functional impact of the transcriptional networks nucleated by TEL-AML1 in different cellular compartments and stages of the disease as well as their vulnerability to therapeutic interventions. Our work begins to extend the analysis of leukaemia to the systems biology level and the knowledge of cellular and molecular targets gained, will further provide a basis for informing novel therapeutic approaches. Key contributions in our programme on leukaemia have been the identification of the cellular targets in which the TEL-AML chimaera first has biological impact (Hong et al 2008, Figure 2) which established the concept of pre-leukaemic stem cells in humans and laid the foundation for a follow up collaborative study which has identified genetic heterogeneity in the stem cell compartment of childhood ALL (Andersen et al 2011). We are now developing these ideas in the settings of childhood AML and initiating a new programme of study in lymphoma led by Rajeev Gupta.
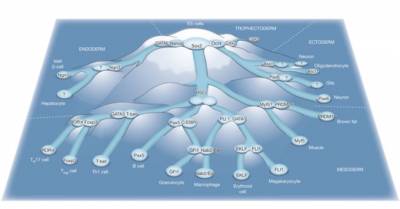
Figure 1: Transcription factor cross-antagonisms in a cascading landscape of unstable and stable cell states. The territory, represented as a mountain range, depicts all possible solutions of a single regulatory network that specifies cell identity. Robust network states correspond to stably differentiated cell types (deep basins in the low-lying plains) whereas unstable solutions correspond to ridges and slopes in the landscape. The latter are only fleetingly occupied during development and thus unlikely to correspond to observable cell types. The route between pluripotent and fully differentiated network states is punctuated by a series of metastable states corresponding to progenitors characterized by the cross-antagonistic interaction of competing lineage-affiliated transcription factors. Within these goggle-shaped ‘binary attractors’ transcriptional networks fluctuate between lineage-biased states before exit either into a stable attractor corresponding to a developmental endpoint, or into a subsequent metastable attractor where a secondary lineage decision is taken. In this model the sequential establishment and resolution of transcription factor cross-antagonisms is a driving force in lineage specification. However, such a mechanism might not apply to earlier intermediates, which may only be partly restricted and do not necessarily commit through simple binary decisions. The intermediates and paths depicted may not be exclusive or obligatory transit points, but rather represent the most favoured possibilities. Although all the transcription factors shown have been experimentally demonstrated to possess lineage-instructive capacity, their precise mechanism of action or the identity of a presumed antagonistic partner (indicated with a question mark) is not known. ES cells, embryonic stem cells; HSCs, haematopoietic stem cells.
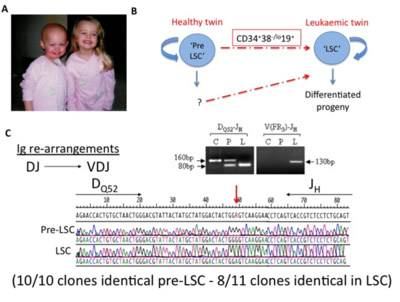
Figure 2: Cell hierarchies and lineal relationships in an evolving TEL-AML1 clone in monochorionic twins. A)The monochorionic twins pictured were naturally conceived. The index child (left) presented at the age of 2 years with TEL-AML1-associated pre-B ALL. The second twin has always been clinically well but her peripheral blood contains a small proportion of 19+ cells with the TEL-AML1 fusion. B)Predicted lineal relationships of the 34+38–/lo19+ populations seen in both twins which was explored through analysis of IgH gene rearrangements: C) DQ52-JH (heavy-chain joining region) (left) and VDJ (right) analyses for control Kasumi cells (C) and 34+38–/lo19+ populations from healthy (P) and leukaemic (L) twin. Arrows indicate 160 bp (germline) and 80 bp (rearranged) segments. Representative sequence analyses of individual cloned DQ52-JH PCR products from healthy and leukaemic twins are also shown. Long arrows indicate oligonucleotide primers, short arrow indicates a G-A transversion suggestive of somatic hypermutation. A minority of (3 out of 11) clones from the 334+38–/lo19+ cells of the leukaemic twin were different from each other and those of her sibling, which may suggest ongoing DJ recombination (see Hong et al 2008 for further details).
 Close
Close


