Group Leader: Dr Suzana Hadjur
Wellcome Trust Senior Research Fellow
Research
Spatial and temporal control of gene expression is essential for the development of complex multicellular organisms. Inappropriate gene activity leads to developmental abnormalities and cancer. My research is focused on understanding the fundamental mechanisms controlling gene expression in mammalian cells.
We are investigating two related questions:
- How does three-dimensional chromosome architecture influence gene expression?
- How do the insulator proteins, cohesin and CTCF, affect cell-specific gene programmes during normal and cancer development?
The eukaryotic genome is organized in three-dimensional (3D) nuclear space in a specific manner that reflects its underlying cell state. With the advent of molecular methods designed to ‘capture’ chromosomes in their native 3D conformations, our understanding of the relationship between chromosome structure and genome function is rapidly changing. Recent studies have revealed that chromosomes are composed of a hierarchical series of chromatin loops. These loops organize large complex chromosomes into discrete, manageable compartments, which are thought to spatially restrict regulatory elements to their target genes, promoting their physical interaction. Understanding the molecular basis of this regulatory architecture will transform our ability to build reliable models of gene regulation and is a major focus of the work in my laboratory.
Cohesin is an ancient and essential protein complex with fundamental roles in chromosome topology. Defects in cohesin and its regulators are increasingly recognized as being responsible for chromosome mis-segregation in human cancers and are the cause of developmental disorders, thought to result from widespread gene deregulation. Cohesin complexes are required for sister chromatid cohesion during mitosis, DNA repair and also function in gene regulation. Cohesin regulates gene activity primarily through the formation/stabilization of chromatin loops. Cohesin-anchored chromatin loops are widely distributed throughout the genome, creating an extensive network of long-range contacts and thereby defining global chromosome topology. Thus, cohesin is a critical component of regulatory architecture and is key to our understanding of how topological configurations of chromatin influence gene expression and consequently cell fate.
We use brain stem cells and their post-mitotic progeny as our model systems. We take a multi-disciplinary approach that combines high throughput experimental and genomics methods and computational analyses (Hi-C, 4C-seq, ChIP-seq) with genetic tools and single cell microscopy to examine how cohesin-mediated nuclear architecture regulates gene expression underlying cell states.
Research highlights include 1) discovery that the mitotic regulator cohesin can also regulate gene expression; 2) discovery that cohesin facilitates communication between distal DNA elements to regulate gene activity; 3) development of high-throughput methods for mapping cohesin-mediated chromatin loops genome wide; 4) cohesin and CTCF anchor chromatin loops throughout the genome at multiple scales; 5) the orientation of the CTCF motif underlies specificity and directionality of long-range chromatin interactions.
We are currently studying how cohesin-mediated nuclear architecture influences the regulation of genes during stem cell differentiation in the mouse. We are developing unique genetic tools and sophisticated systems to build a comprehensive understanding of how topological conformations of chromatin collaborate with transcription factors to drive lineage commitment.
Corruption of cell differentiation is also a hallmark of cancer. Understanding how cancer stem cells develop is of great importance. We are investigating the brain tumour glioblastoma, where mutations in cohesin proteins have been identified. We aim to shed light on cancer development through understanding of the molecular mechanisms underlying control of chromatin structures.
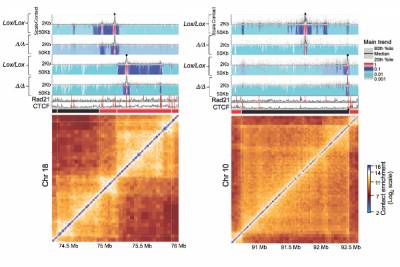
Fig.1 - High-resolution 4C-seq analysis of cohesin/CTCF-mediated interactions. 4C-seq profiles of baits (black spots) anchored at cohesin/CTCF sites in two different genomic regions in control (Lox/Lox) or cohesin- deficient cells (Δ/Δ). Also shown are the ChIP-seq tracks for Rad21 and CTCF as well as the corresponding Hi-C maps. The results demonstrate preferential and asymmetric cohesin-cohesin interactions which are lost in Rad21-deficient cells.
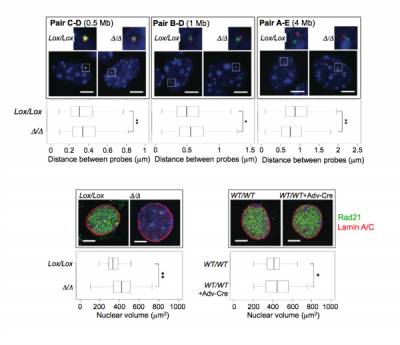
Fig.2 - Cohesin-deficient cells experience global chromosome domain decompaction and increased nuclear volume. Three sets of FISH probes were designed to the borders of chromosomal domains and labelled in red or green. Three-dimensional inter-probe distances in control (Lox/Lox) and cohesin-deficient cells (Δ/Δ) were measured using Volocity software. In the absence of Rad21, domain borders decompacted. Genomic distances between probes are indicated in parentheses. White dotted boxes show the regions that have been zoomed in. (Bottom) Cohesin-deficient nuclei were significantly larger compared to controls. Whiskers and boxes indicate all and 50% of values respectively. Central bold bars represent the median. Scale bar = 5 μm..
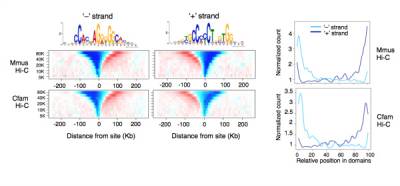
Fig.3 - Oriented CTCF motifs determine the directionality of CTCF-mediated interactions. Average contact insulation analysis in both Mmus and Cfam genomes for conserved CTCF binding sites grouped according to the orientation of CTCFs binding motif. (Right) Genome-wide relative position within chromosomal domains of conserved CTCF sites grouped according to the orientation of their binding motif as above.
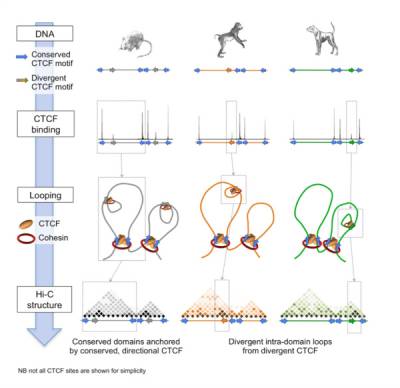
Fig.4 - Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. To explore the mechanisms underlying the evolution of chromosomal domain structures, we compared four mammalian species and revealed a direct link between insulator site divergence and the evolution of chromatin domain structure. The data point to a direct role for CTCF/cohesin in driving structural change in the genome.
Selected publications
- Pezic D, Weeks SL, Hadjur S. (2017) More to cohesin than meets the eye: Complex diversity for fine-tuning of Function. Current Opinion in Genetics and Development 2017 Apr;43:93-100.
- Barrington C, Finn, R, Hadjur S. (2017) Cohesin Biology meets loop extrusion. Chromosome Research 2017 Mar;25(1):51-60.
- Uusküla-Reimand L, Hou H, Samavarchi-Tehrani P, Vietri Rudan M, Liang M, Medina-Rivera A, Mohammed H, Schmidt D, Schwalie P, Young EJ, Reimand J, Hadjur S, Gingras AC, Wilson MD. (2016) Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biology. Aug 31:17(1):182-86
- Vietri-Rudan M, Hadjur S* and Sexton T*. (2016) Detecting spatial chromatin organization by chromosome conformation capture: genome-wide profiling by Hi-C. Methods. Methods in Molecular Biology. Feb 23
- Vietri-Rudan M and Hadjur S. (2015) Genetic tailors - CTCF and cohesin shape the genome during evolution. Trends in Genetics. Nov:31(11):651-60.
- Vietri-Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. (2015) Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Reports. Feb 23:1-14
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri-Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. (2013) Cohesin stabilizes interphase chromosomal architecture. EMBO J. Dec 11; 32(24): 3119-29.
- Martynoga B, Mateo JL, Zhou B, Andersen J, Achimastou A, Urbán N, van den Berg D, Georgopoulou D, Hadjur S, Wittbrodt J, Ettwiller L, Piper M, Gronostajski RM, Guillemot F. (2013) Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. Aug 15: 27(16): 1769-86.
- Sofueva S and Hadjur S. (2012) Cohesin-mediated chromatin interactions – into the third dimension of gene regulation. Briefings in Functional Genomics. May; 11(3): 205-16.
- Hadjur S, Ryan NK, Williams LM, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. Jul 16;460(7253):410-3.
- Parelho V*, Hadjur S*, Spivakov M, Leleu M, Sauer S, Gregson H, Jarmuz A, Webster Z, Nesterova T, Cobb, B, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. Feb 8;132(3):422-33.
*equal contribution
 Close
Close




