The Genomics TTP provides access to state-of-the-art genomics equipment and offers NGS library preparation and genome engineering services.
The Translational Technology Platform (TTP) is supported by experienced specialists who can offer their expertise and support at all stages of the research project. We can help with experimental design, providing technical services, and training to facilitate your research projects requiring genomics and/or genome engineering technologies. You can download the Genomics TTP leaflet here:
We provide comprehensive sequencing services including next-generation sequencing (NGS) library preparation and sequencing using the Illumina NextSeq 550 and MiSeq. We work closely with the Bioinformatics TTP, based at the Bill Lyons Informatics Centre (BLIC), also supported by the CRUK UCL Centre, who can provide help and support with sequencing data analysis for various applications.
The Genomics TTP also closely works with the CRUK City of London Centre Single Cell Genomics Facility to run samples on the 10X Genomics Chromium Controller and Fluidigm C1 platforms and provide a streamlined service from library prep to bioinformatic analysis.
Our genome engineering services include production and use of the latest CRISPR technologies as well as banking and distribution of associated in-house developed CRISPR reagents. We also have the Human and Mouse Open Biosystems GIPZ shRNAmir lentiviral libraries, which can be purchased as individual hairpins or as pooled libraries for multiplexing screening. The TTP provides support in assay design and use of the RNAi libraries collections.
We currently offer a MiSeq service for sequencing edited cell populations or single cell clones. Please contact us for more information about this service.
Equipment
There are a number of genomics platforms available within the TTP. We are happy to undertake projects for the research groups based within the Cancer Institute and the wider CRUK UCL Centre and also provide training to researchers who would like to use our equipment.
Please contact ci.genomics@ucl.ac.uk to discuss your project and training requirements and costs. Following appropriate training, you will be given access to the relevant booking calendars on the Clustermarket Bookkit system.
- MiSeq
Location: Lab 228
The Illumina MiSeq is a highly automated benchtop sequencing system suitable for a wide range of applications such as Small genomes sequencing, Targeted resequencing, Small RNA sequencing, Metagenome sequencing, de novo transcriptome sequencing and more. It is fast and cost-effective with short sequencing run times and long reads while maintaining high data quality. A standard MiSeq run is routinely capable of delivering between 1-25 million reads dependent on the needs of the experiment and kit selection.
The current MiSeq kits that are available include Nano, Nicro and Standard kits that can output up to 1, 4 and 25 million reads respectively.
Due to the economical nature of MiSeq Nano kits, we often recommend that these kits be used to QC libraries before running them on the Illumina NextSeq, or outside the institute on Illumina NovaSeq instruments.
- NextSeq 550
The NextSeq 550 is housed within Barclay House, GOSH so only TTP staff have direct access. Please contact us for processing of your samples.
The Illumina NextSeq 550 System combines industry standard Illumina instrument technologies and customisable output with sequencing and array capabilities. Users can run transcriptome, targeted resequencing, genotyping, and other applications all on one platform. The NextSeq can be configured to run in either mid or high output mode. The current range of kits that are available come in a range of sizes which makes it possible to tailor the sequencing requirements to each individual experiment.
The current mid output kits include 150 and 300 cycle options that can output up to 130 million single ended, or 280 million paired end reads.
The current High output kits include 75, 150 and 300 cycle options that can generate up to 400 million single ended, or 800 million paired end reads.
- nCounter SPRINT System
Location: Lab 303B
The nCounter SPRINT system from Nanostring Technologies utilizes a digital barcode technology for direct multiplexed measurement of nucleic acids with high levels of precision and sensitivity. The technology uses fluorescent labelled reporter probes called ‘codesets’ that bind to the target molecules and single molecule imaging to detect and count hundreds of molecules in a single reaction (<1 copy per cell). Pre-designed Codesets are available for mRNA, DNA and miRNA and custom Codesets can be designed. The major advantage of the technology is the ability to analyse RNA samples of poor quality such as FFPE (formalin-fixed, paraffin-embedded) samples.
Please note: this equipment is not currently covered by a service contract but is available for use.
- QuantStudio 5 Real-Time PCR System
Location: Lab 303C
The QuantStudio 5 Real-Time PCR System is designed for users who need a high performance instrument with features for maximum experiment control. The instrument as well as the Design and Analysis software are very easy to use.
- Eppendorf MasterCycler RealPlex qPCR Instruments
Location: Lab 303C
We have three Mastercycler RealPlex instruments from Eppendorf which are compact, easy to use and flexible platforms to obtain accurate, consistent and reproducible results. The instruments are compatible with plates and reagents from different companies which make these an ideal and flexible platform to use.
- Agilent 2100 Bioanalyzer Instrument
Location: Lab 303C
The Agilent 2100 Bioanalyzer provides sizing, quantification and purity assessments for DNA, RNA and protein samples. Kits for the following assays are kept in stock by the Facility – RNA nano, DNA 1000 and DNA High Sensitivity.
- Qubit 3.0 Fluorometer
Location: Lab 228
The Qubit 3.0 Fluorometer is an extremely simple to use benchtop fluorometer that can be used for the quantitation of DNA, RNA, microRNA and protein using the highly sensitive and accurate fluorescence-based Qubit quantitation assays. Use of dyes that are selective for dsDNA, RNA or protein minimizes the effects of contaminants in the sample. Data can be easily exported to a USB drive.
- Tapestation 2200
There is a Tapestation available in the Pathology Translational Technology Platform. Please see the Pathology Translational Technology Platform page for further details and booking.
Genomics Services
Please contact ci.genomics@ucl.ac.uk to request the order form for submitting your samples.
- Sequencing
The TTP provides sequencing services if you wish to prepare your own libraries. We have access to Illumina MiSeq and NextSeq 550 instruments to cater for both small and large genomics projects. Based on your study requirements, we can advise on relevant sequencing kits and consumables that will be needed for your sequencing runs.
- NGS library preparation
We offer library preparation services for mRNA-Seq and Exome-Seq. As part of our service, we will also quantify libraries and check their quality.
- Methylation profiling
The Illumina Infinium MethylationEPIC BeadChip allows profiling of over 850,000 methylation sites at single nucleotide resolution (> 90% of the original CpG sites from Human Methylation 450BeadChip plus an additional 350,000 CpG sites in enhancer regions). Multiple samples can be processed in parallel to provide a broad and comprehensive view of the methylome.
We facilitate research projects requiring methylation profiling by performing bisulfite conversion, quality control and liaising with the UCL Genomics Facility to get those samples processed on the Infinium MethylationEPIC BeadChips.
We also process FFPE samples for methylation studies by including extra QC and DNA restore steps to the standard protocol.
Single Cell Genomics
The CRUK City of London Centre Single Cell Genomics Facility provide a streamlined service from library prep to bioinformatic analysis. We can run samples on the 10X Genomics Chromium Controller and Fluidigm C1 platforms and can run single-cell RNA-seq, scATAC-seq and CITE-seq experiments and are open to discussions about adding other methods you may be interested in.
Please see more details on the Single Cell Genomics Facility website.
Genome Engineering Services
Please note that these services are available only to members of UCL
- Open Biosystems pGIPZ lentiviral shRNAmir RNAi Libraries
We currently have the following shRNA libraries:
- Open Biosystems pGIPZ lentiviral shRNAmir library- HUMAN
- Open Biosystems pGIPZ lentiviral shRNAmir library- MOUSE
These libraries can be purchased as individual hairpins or as pooled libraries for multiplex screening.
You should contact your safety officer prior to beginning this work to ensure you have all the appropriate risk assessments and safety precautions in place. Sonia Buckingham is the Health and Safety Lead for the UCL Cancer Institute: s.buckingham@ucl.ac.uk
Open Biosystems pGIPZ shRNAmir library details
We have the Human and Mouse Open Biosystems GIPZ shRNAmir lentiviral libraries. Both libraries contain over 100,000 active clones, each containing a unique hairpin to target knockdown of genes within the Human and Mouse genomes respectively.
shRNAmir
The design of the hairpins contained within the GIPZ vector is based on the endogenous mir-30. By mimicking an endogenous RNA, the GIPZ hairpin is efficiently processed in vivo allowing for a more effective knockdown of your gene of interest.
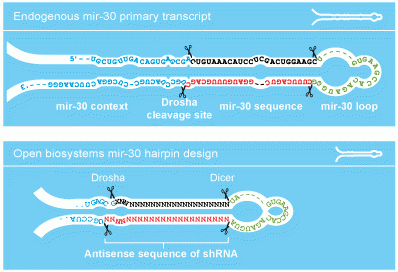
GIPZ vector design
The GIPZ lenti-vector contains various features to allow effective expression, tracking and versatility of your hairpins. These features include:
- CMV (Pol II) promoter
- Turbo GFP to mark shRNAmir expression
- Puromycin selectable marker
- Lentiviral vector to allow successful transduction of primary, non-dividing and other difficult to transduce cells
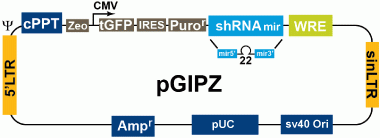
Further information
- This plasmid map can be opened using Snapgene Viewer which is free software and can be downloaded
- See GE website for further information about libraries
- GIPZ vector map:
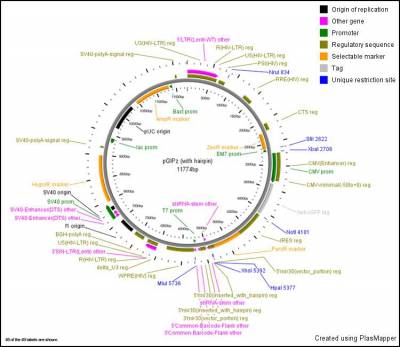
Ordering
Individual hairpin clones
These GIPZ hairpins and any derivatives (viruses, cell lines, portions of the GIPZ vector cloned into other vectors etc.) must not be shared or transferred to institutes outside UCL. They must remain and be used solely within UCL laboratories.
You should contact your safety officer prior to beginning work with GIPZ hairpins to ensure you have all the appropriate risk assessments and safety precautions in place.
Formats
Individual hairpins come as bacterial agar stab cultures. You will need to re-streak the bacteria from the surface of the culture onto an LB-Amp plate, pick a single colony, grow, prepare and purify your plasmid DNA using a kit and sequence the DNA to make sure the sequence is correct. The DNA is then ready for transfection.
Controls
Negative:
- Empty GIPZ vector
- Non-silencing (scrambled) (22mer: aTCTCGCTTGGGCGAGAGTAAG)
Positive:
- GAPDH (22mer: cCCTCATTTCCTGGTATGACAA)
- EG5 (22mer: cGGCCATGCTAGAAGTACATAA) Targets Human but not Mouse
Controls are the same price as the shRNA hairpins and can be ordered by contacting ci.genomics@ucl.ac.uk to request the order form.
UCL Consortium prices
This includes members of the UCL Cancer Institute, Institute of Child Health, Institute of Neurology and the Division of Infection and Immunity.
Bacterial Stab Cultures: £15.00
Ordering clones
Instructions:
- Contact ci.genomics@ucl.ac.uk to request the order form
- Go to the GE website and follow the GIPZ Ordering Instructions to search for available hairpins. We have all of the Human and Mouse GIPZ hairpins within our collection starting with the codes V2LHS, V3LHS, V2LMM or V3LMM.
- Take note of the CloneID for the hairpins you would like
- If you are not from within the Cancer Institute, please make sure that you fill in your project details and Finance contact within your institute on the order form to enable us to generate an eIDT. Refer to the price list above, if you are unsure please contact us at ci.genomics@ucl.ac.uk
- Complete the ordering form:
- Make sure you enter your project code (CI as well as non CI users)
- Indicate whether you would like any controls and what format you would like
- List the the CloneID of the hairpins you are after. These codes will start with V2LHS, V3LHS, V2LMM or V3LMM
- Email your completed form to ci.genomics@ucl.ac.uk. We will contact you to let you know your order has been received and to arrange collection. All clones are collected from the UCL Cancer Institute, Paul O'Gorman Building.
Sequencing your hairpins
We highly recommend sequencing your hairpins before starting work with them.
GIPZ sequencing primer: 5' - GCATTAAAGCAGCGTATC - 3' Melting Temp: 52.7c The binding site for this primer is base 5820-5842 and runs in reverse complement direction.
- CRISPR sequencing service
Our Facility also runs a highly cost-effective targeted MiSeq CRISPR amplicon sequencing service to sequence edited cell populations or single cell clones. The CRISPR sequencing service was developed 'in house' to provide deep sequencing of amplicons derived from CRISPR-targeted genomic loci. The advantage of this method is the ability to identify and quantitate the type of insertions and deletions (indels) in any given sample, at the sequence level. Please contact us on ci.genomics@ucl.ac.uk for more information and associated costs.
- CRISPR Reagents
We provide support in the design, production and use of the latest CRISPR technologies as well banking and distribution of associated in-house developed CRISPR reagents.
CRISPR reagents can be ordered directly from addgene. Please contact ci.genomics@ucl.ac.uk for more information and protocols for CRISPR reagents, including CRISPR/Cas9 Plasmids.
CRISPR/Cas9
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated systems
The CRISPR/Cas9 system is currently the most popular genome engineering technology available. It is a DNA binding complex based on a bacterial immune system that can be easily customised to target a specific sequence of DNA.
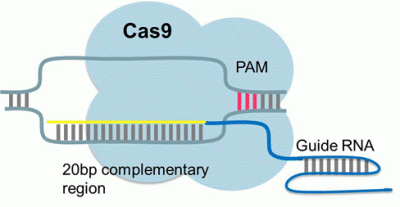
The system utilizes two components 1) a short guide RNA (gRNA) to recognise and bind target DNA and 2) a CRISPR associated (Cas9) endonuclease. When expressed together, the gRNA and Cas9 form an active endonuclease. The gRNA binds to the target sequence guiding a wildtype Cas9 to cleave the double stranded DNA in a site specific manner and resulting in DNA editing via DNA repair pathways.
The CRISPR/Cas9 system is very versatile and many Cas9 variations exist. This includes nuclease deficient Cas9, which allows this system to be used as a DNA activation, repression, localization or purification tool and so on.
The CRISPR/Cas9 system is a rapidly changing and advancing technology. New CRISPR/Cas9 reagents are constantly being developed, expanding even further upon the capabilities of this system. The CAGE facility aims to help researchers to utilise this system.
We currently have a number of CRISPR/Cas9 reagents that can be ordered from the TTP.
- Lentiviral Production
We can provide high titre, second and third generation lentivirus directly from the Open Biosystems Human and Mouse whole genome shRNA libraries (for information on the libraries, please see the link above for RNAi Libraries). We also offer a custom lentiviral packaging service that is dependent on the provision of a suitable second or third generation lentiviral vector by the researcher. Please contact us on ci.genomics@ucl.ac.uk for more information and associated costs.
 Close
Close








