Stem cells are undifferentiated cells with the unique ability to self-renew - that is, to maintain their stem cell status indefinitely as they divide. In some cases they can also be multipotent (ie. they can give rise to many different types of specialised cells). Stem cells play an important role in the development of the embryo but are also present in the adult. Considerable effort is currently being invested worldwide to harness for many different clinical applications, including therapies for many diseases and as a means of testing the effectiveness of pharmaceutical compounds in the laboratory. We are currently undertaking three research projects on stem cells:
1. Endogenous stem cells in the embryo
Many years ago we discovered that when a single cell in Hensen's node of a chick embryo is labelled with a fluorescent dye, its progeny spread along the axis of the embryo. Interestingly, if this cell contributed to somites, there were clusters of labelled descendants at intervals of 6-8 somites, while if the cell contributed to notochord the clusters were 2-4 somites apart. Occasionally we labelled a cell which contributed both to somites and to notochord, in which case the descendants were spaced 6-8 somites apart in the somites and 2-4 somites apart in the notochord. Strikingly in all cases a single labelled cell could still be seen in the remnants of the node (Selleck & Stern, 1991; Selleck & Stern 1992).
These findings strongly suggest the existence of resident populations of stem cells in Hensen's node - there are stem cells for somites (in fact they only contribute to the medial halves of the somites) and stem cells for notochord, and there are parent cells that can give rise to both. One pair of somites forms every 90 minutes in the chick, therefore a spacing of 6-8 somites corresponds to about 10 hours, which was demonstrated to be the cell cycle time of somite precursor cells (Primmett et al., 1989). We calculated that only 32 somite precursor stem cells on each side of the embryo are necessary to generate all the cells that will form part of the medial somite (a newly-formed somite contains about 1,000 cells, so the 32 precursors would divide 4 times to give rise to 512 cells in the medial halves of the somites).
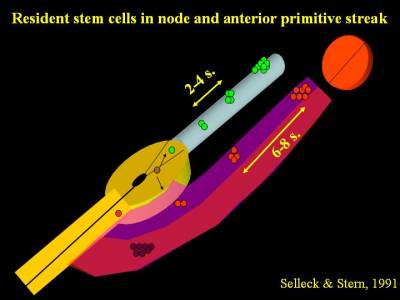
This was the first demonstration of endogenous stem cells residing in the normal embryo during development. Since then, findings consistent with these have been made in the mouse by the groups of Rosa Beddington, Val Wilson and Jean-Francois Nicolas and in the chick by Isabel Palmeirim and it is now generally accepted that a resident population of stem cells is likely to account for how the embryo grows in a head-to-tail direction during development. These findings are consistent with the idea that the resident stem cells are true self-renewing stem cells, which divide asymmetrically to generate one daughter that enters the mesoderm (as either a somite or a notochord precursor) while the other daughter remains in the node until its next division. The problem is that the calculated 32 cells are hidden among some 2,000 other cells in the node - a real needle in a haystack problem. How can we find them reliably, so that we can study them further? We are currently using 2-photon microscopy to identify and follow the divisions of cells in the node in time-lapse movies and also studying the expression of genes found to be required for asymmetric cell division in other systems.
2. Genes regulating multipotency and commitment and their role in cancer
Formally there are only about 7 distinct signalling pathways through which cells can communicate with one another. One of the great mysteries of development is how can the embryo generate such a huge diversity of cell types and cell behaviours with so few pathways, since even all possible combinations of 7 pathways is not enough to account uniquely for all the cell types in the body. The answer must lie in the timing of how these pathways are deployed - cells seem to accumulate a "history" of what signals they have received over time, and therefore the same signal will have different effects depending on what other signals the receiving cell has experienced at earlier stages of development. Integration of this "memory" must be done, at least in part, at the level of regulatory regions of genes. These considerations mean that if we are trying to manipulate stem cells in a culture dish, it will be impossible to obtain all possible cell types just by exposing the cells to single, or combinations of, extracellular signalling proteins. How then can we hope to manipulate their fate?
Our research on neural induction has uncovered several genes which act to regulate the timing of how cells interpret signals. One of them is the transcription factor Churchill is induced relatively slowly by FGF signalling and thus acts as a switch between early (mesoderm-inducing) and later (neural-inducing) activities of the FGF pathway (Sheng et al., 2003). Several of these genes appear to act at the level of chromatin, regulating its availability for transcription of targets which in turn control cell fate. We are now investigating whether genes that act as switches, causing cells to respond differently to the same signals depending on the lineage and signalling history of the cells, also regulate commitment of stem cells to certain fates. One approach to understand how these switches work is by trying to reconstitute the whole cell decision system in embryonic stem cells (ES cells) (see below). The aim is to direct ES cells to acquire particular fates (for example neural) by manipulating their expression of these switches rather than just exposing them to different external factors - we are now trying to use the information from our neural induction project to direct ES cells to adopt a neural fate in a predictable way. One intriguing possibility, since cancer is a disease in which cells fail to differentiate and instead continue to proliferate uncontrollably, is that the same genes that control multipotency and commitment during early development have gone awry in certain types of cancer.
We are currently collaborating with the Institute of Molecular Oncology in Milano (Italy) by analysing large tissue arrays of human tumours for the levels of expression of the commitment genes, to determine whether there is a correlation between expression of these genes with cancer prognosis or tumour invasiveness.
3. Chicken embryonic stem cells (ES cells)
Embryonic stem cells have been available from a variety of mammalian species for some time (initially from mouse, then rat and more recently many other mammals). Since they can contribute to all somatic lineages as well as to the germ line, and since they can be genetically manipulated by homologous recombination, ES cells have been exploited as a very effective means of generating transgenic mice. However until very recently it had been impossible to generate ES cells, which can be kept in culture indefinitely while retaining their pluripotency, from non-mammalian vertebrates. Over the last few years, groups in North Carolina (Jim Pettite), San Francisco (Robert Etches) and Lyon (Jacques Samarut and Bertrand Pain) have succeeded in generating cell lines from very early chick blastoderms. These cells can be kept indefinitely in culture and seem to be able to contribute to all somatic lineages (although no success has yet been obtained in terms of a contribution to the germ line).
We have several such cell lines in the laboratory and can also generate new lines. These can be manipulated genetically and are susceptible to homologous recombination, which makes them a fantastic tool to study their ability to contribute to different lineages after being manipulated. We are using these cells to reconstruct the decision of early embryonic cells to become neural based on the results of our neural induction project. We are also transplanting normal and genetically manipulated ES cells back into normal embryos and testing their ability to contribute to neural or other tissues based on the manipulation performed.
 Close
Close

