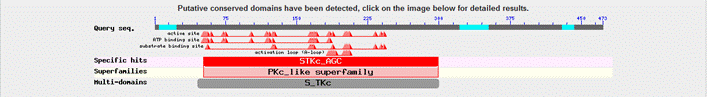
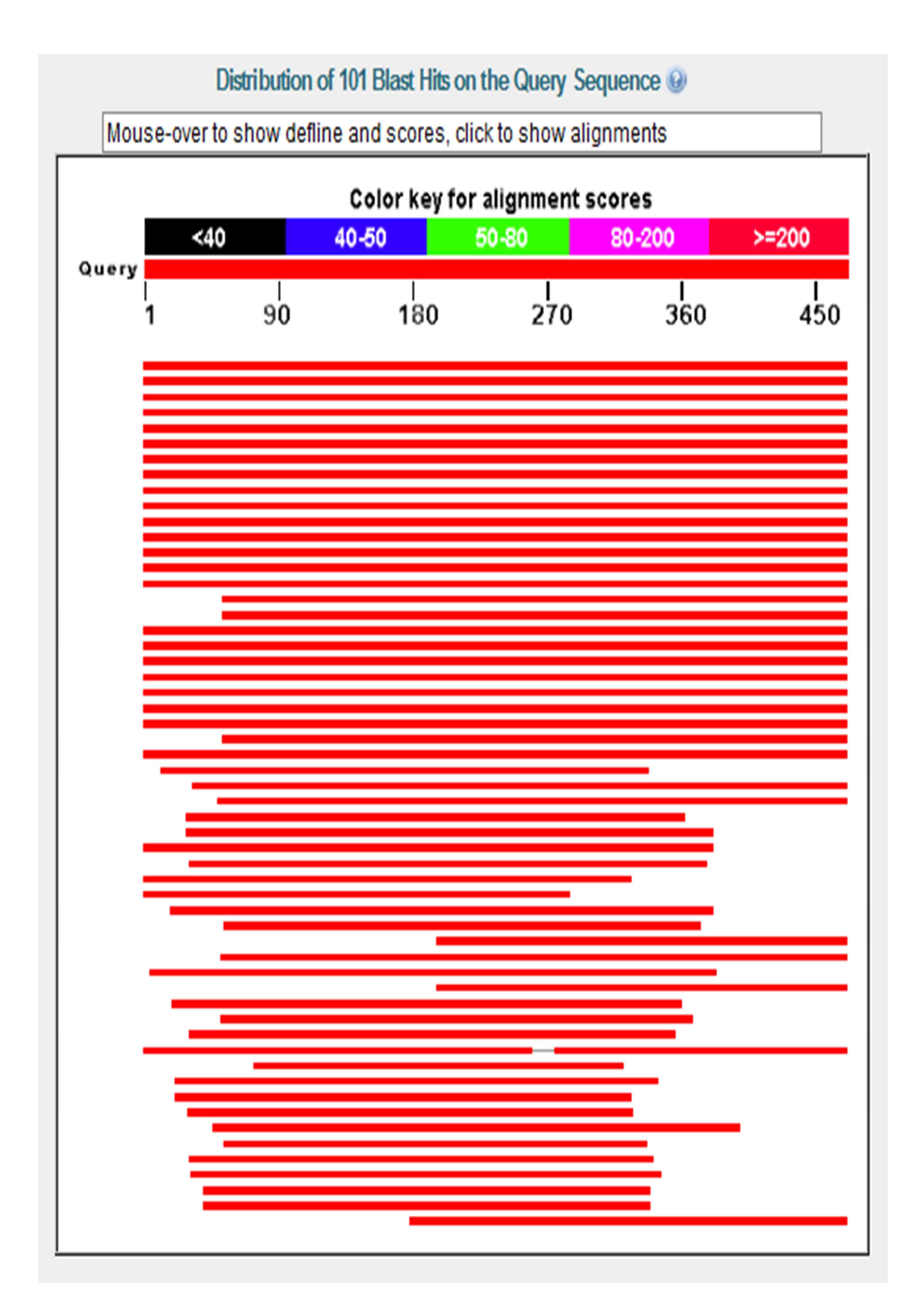
Basic Local Alignment Search Tool (BLAST)
A BLAST search was done using the human CaMKIVα isoform sequence (our representative protein). The results do not seem to be particularly incisive, as many of the “BLAST hit” proteins are not clearly labelled. Many CaMKIV proteins that displays a high degree of homology to the human CaMKIVα are based on predicted models, which could suggest that CaMKIV might be a protein that is currently being extensive studied by many molecular scientists. As can be seen from the diagram, we have ordered our blast results on the basis of total score (which is calculated using the bit score of HSPs - high scoring segmental pairs). A higher total score therefore indicate a more extensive stastical alignment between the representative protein sequence (the one that is entered for BLAST) and the corresponding protein sequence, this in turn suggests that there is a greater degree of sequence homology between the two proteins. From the blast results we can immediately see that the total score (abbreviated to TS) of mouse, rat and cow, which are all mammals (TS of 727, 710 and 705 respectively), is much higher than those from, for example, amphibian and zebrafish (TS of 622 and 608 respectively). This indicate a significant extent of evolutionary conservation of CaMKIV within different related species. This result also supports the evolutionary relationship of CaMKIV between different species from our phylogenetic tree. The result also indicates CaMKIV from some species such as macaques (TS = 842) are not yet officially sequence, as only predicted values are given. The E (expected) values highlights the probability of obtaining a specific protein sequence by pure chance, this in turn highlights the significance and reliablility of the obtained results. Although our BLAST search has an overall E value ranging between 0 and 1 × e ^ -88 (which are spread across 3 'pages', the E value that falls within our area of interest (the results that includes CaMKIV sequences - which includes all results up to the end of the second 'page') actually ranges between 0 and 3 × e ^ -92). This indicates that all results up to that specified point is extremely significant and reliable. This BLAST was carried out using the blastp (protein blast) search on the BLAST section of the NCBI website. For more details regarding how to analysis a BLAST result, please click here. |