Recent years have seen a vast increase in the numbers of protein sequences, with several thousand genomes now available, each of these typically containing thousands of genes. How these genes work in combination to perform complex essential functions in cells is one of the main challenges that our lab and others are working on. A number of algorithms have been developed within the group to build and make networks of gene associations to help with this task.
CODA
The group have developed various methods for predicting functional interactions based on domain fusion events. The CODA method uses multiple genomes to predict functional associations based on detected domain fusion events. Such approaches can be applied computationally to all genomes.
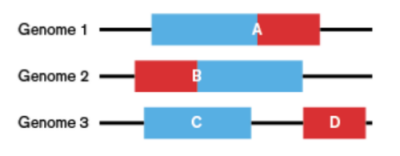
Figure illustrating the principal of CODA whereby domains (rea and blue sections) are found fused in some genomes (1 and 2) genomes but found on separate chains in a query genome 3.
Phylotuner
Loss and gain of functions and whole pathways has commonly occurred across of the tree of life. Now that sufficient genomes are available, functional associations between genes can be inferred by looking for co-occurrences of gene family gain and loss. Phylotuner is unique in that it makes use of domain based families and applies novel metrics to allow its use in higher eukaryotes with large gene families.
As more genomes become available such methods are likely to become more powerful.
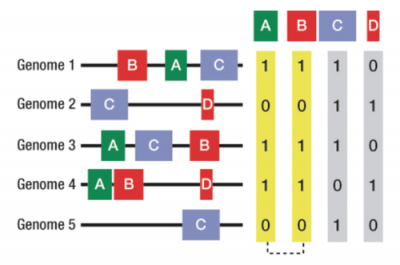
Figure showing co-occurrence of domain families A and B across multiple genomes implying a functional association.
HIPPO
The group works on many different organisms and as such we have come up with a method to transfer interactions between them. These inherited interactions enable us to carry out network based analysis in organisms for which only a few experimental interactions are characterised.
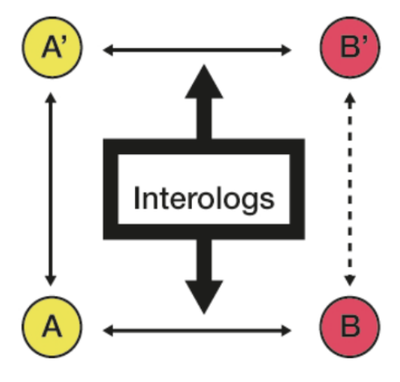
Figure experimental interactions between proteins A and B and inherited by orthology to infer the interaction A'-B'.
Fun-L
Fun-L (http://funl.org) makes use of a number of protein interaction data sources to identify sets of proteins likely to be working together in the same biological processes. The networks are first transformed into kernel matrices and combined. Developing these kernel based methods is an active area of research in the lab, and we are collaborating with Prof. John Shawe-Taylor from Computer Science in this task. We have successfully applied these methods to better understand the mitotic chromosome condensation process.
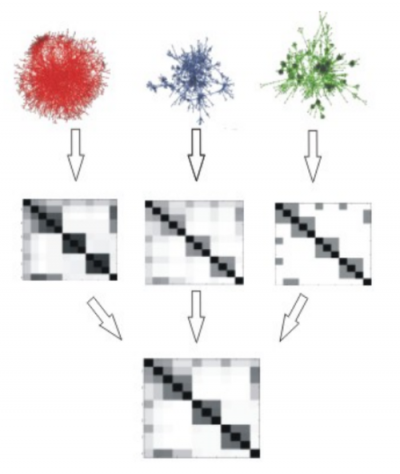
Figure describing Fun-L showing how different source protein interaction networks on the left are converted into individual kernel matrices middle and finally combined into a single integrated matrix.
Network Dynamics
The group is also interested in studying network alterations and dynamics through combining multiple experimental datasets. For example we are using proteomics datasets to study network rewiring involved in the stress response and in embryogenesis.
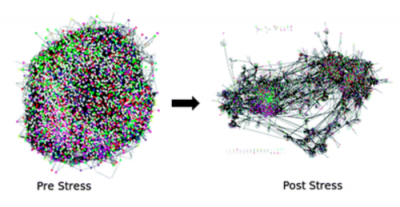
Figure of network undergoing rearrangements on Stress taken from Molecular Biosystems Publication http://pubs.rsc.org/en/Content/ArticleLanding/2013/MB/c3mb25548d#!divAbstract
 Close
Close

