New publication in Open Biology for Acton Lab and colleagues
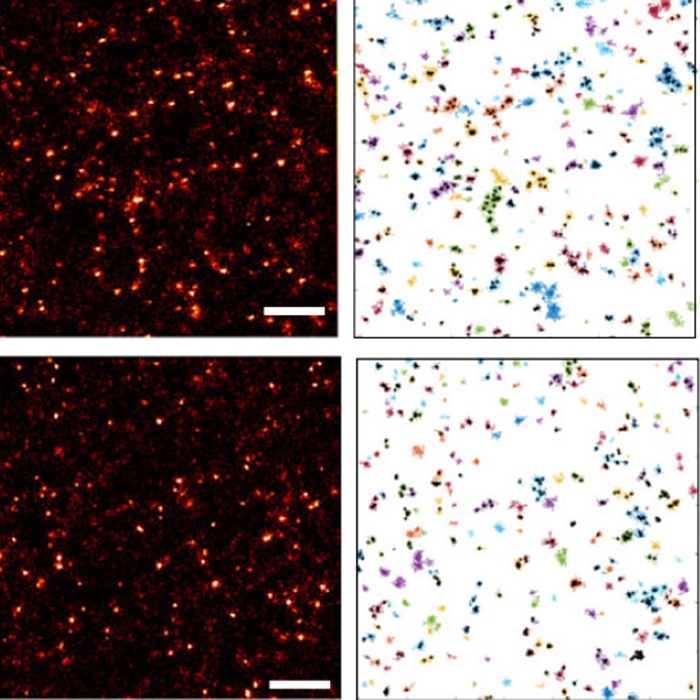
In a newly published study in Open Biology, the Acton and Simoncelli Labs (UCL London Centre for Nanotechnology) used DNA-PAINT, a quantitative single molecule super-resolution technique, to visualize and quantify how podoplanin clustering is regulated in the plasma membrane of fibroblasctic reticular cells.
Abstract:
Upon initial immune challenge, dendritic cells (DCs) migrate to lymph nodes and interact with fibroblastic reticular cells (FRCs) via C-type lectin-like receptor 2 (CLEC-2). CLEC-2 binds to the membrane glycoprotein podoplanin (PDPN) on FRCs, inhibiting actomyosin contractility through the FRC network and permitting lymph node expansion. The hyaluronic acid receptor CD44 is known to be required for FRCs to respond to DCs but the mechanism of action is not fully elucidated. Here, we use DNA-PAINT, a quantitative single molecule super-resolution technique, to visualize and quantify how PDPN clustering is regulated in the plasma membrane of FRCs. Our results indicate that CLEC-2 interaction leads to the formation of large PDPN clusters (i.e. more than 12 proteins per cluster) in a CD44-dependent manner. These results suggest that CD44 expression is required to stabilize large pools of PDPN at the membrane of FRCs upon CLEC-2 interaction, revealing the molecular mechanism through which CD44 facilitates cellular crosstalk between FRCs and DCs.
 Close
Close