Taki Nishimura on his recent publication in Molecular Cell
We've asked Taki Nishimura to tell us about his recent publication in Molecular Cell.
What are you trying to understand?
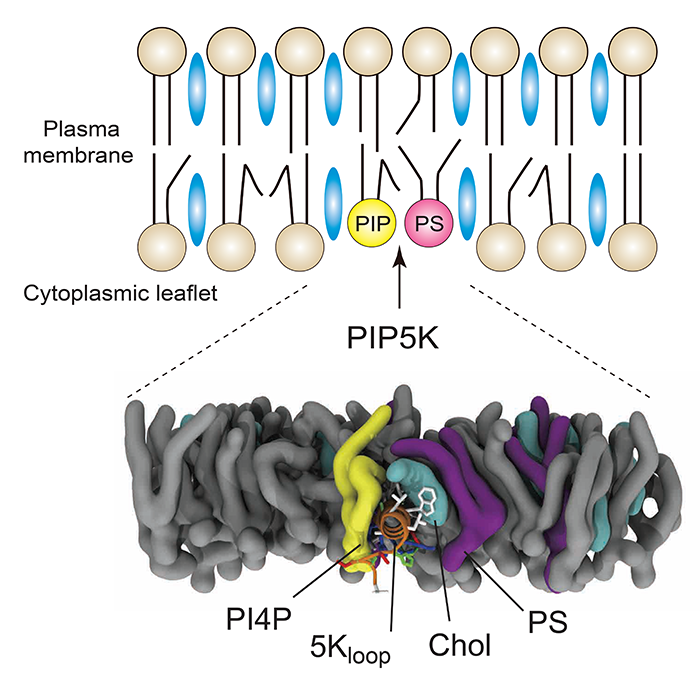
We wanted to gain a better understanding of the architecture of the cell membrane, also known as the plasma membrane. Our study focused on membrane lipid organisation that has been incredibly difficult to study because there are few tools currently available. Specifically, we studied how a well-known signalling lipid called phosphatidylinositol (4,5)-bisphosphate, or PI(4,5)P2, is regulated in cells. PI(4,5)P2 is thought to serve as a ‘signpost’ for plasma membrane identity and it is essential for vital cellular functions and events taking place at the plasma membrane during cell growth and division. So far, physiological roles of PI(4,5)P2 have been intensely studied, but the regulation machinery of PI(4,5)P2 synthesis had remained unclear. PI(4,5)P2 is generated by a lipid kinase and we reasoned that this enzyme might be directly regulated by lipids in the plasma membrane. This was the starting point for our study and this turned out to be the case!
What model system/techniques did you use?
This was a truly inter-disciplinary and collaborative study. We took advantage of and developed new quantitative in vivo, in vitro, and in silico approaches. The in vivo cell biology experiments included quantitative microscopy using novel probes that we designed here in the LMCB as well as state-of-the-art lipidomics with our collaborators in Japan and Germany. In order to validate our working hypothesis based on our in vivo analyses, we performed in vitro biochemical and biophysical experiments. Furthermore, we ran molecular dynamics simulations to build models of nanoscale lipid environments as well as interactions between membrane lipids and the kinase by collaborating with Dr. Hummer’s group at the Max Planck Institute. Our in vivo experiments utilised yeast cells as a model system and our in vitro and in silico investigations were based on a metazoan lipid kinase. Amazingly, the regulatory mechanisms and principles revealed in our study are conserved across species from a simple single-cellular eukaryote to vertebrates!
What has been your most exciting discovery?
A surprising and exciting finding that emerged from our study is that an internal cellular compartment called the endoplasmic reticulum (ER) plays a central role in controlling plasma membrane (PM) composition. Lipids are generated in the ER and then transferred to other organelle compartments by lipid transfer proteins (LTPs). In our study, we found that lipid exchange between the ER and the PM is required for efficient PI(4,5)P2 synthesis. LTPs create an unique lipid environment in the PM that is critical for enzymatic activity of PIP5K, the enzyme that generates PI(4,5)P2. In addition, we characterized a lipid-sensing motif highly conserved in PIP5Ks. The motif has an ability to recognize an environment containing unsaturated phosphatidylserine (PS) and phosphatidylinositol 4-phosphate (PI4P) and sterols that are enriched at the PM. I think that this is the most exciting and important point in our study. Probably, the sensing machinery is a major reason why PI(4,5)P2 synthesis mainly occurs at the plasma membrane rather than other membrane compartments in the cell. It is due to an enrichment of PS, sterol, and PI4P (set up by LTP-mediated exchange) that is a hallmark of the PM. Furthermore, the result of the simulation analysis is amazing! It shows a molecular view of lipids and the sensing motif revealing how the lipid-sensing motif can recognize specific lipid species. Lipid environments and their detection machinery seem to be more complex than we ever thought.
Why is your research important?
Our study has proposed new concepts about membrane biology. For example, it had been unclear why LTPs transfer lipids between organelles, as lipids are also transported by vesicles. Our study provides a good answer to this question. LTPs are able to exchange lipids and efficiently generate unique membrane environments that specify certain activities, such as PI(4,5)P2 synthesis. Vesicular transport may not be capable of generating this precise specificity. In short, a key role of some LTPs might be to stimulate enzymatic reactions by generating membrane environments that promote their activity. Another point is that our study highlights a key role for the ER in directing PM events. Our study implicated sites of contact between the ER and PM, and the formation of membrane contact sites might be of general importance for organelle membrane biogenesis and homeostasis throughout the cell.
What’s next?
Although we examined a regulatory mechanism for PIP5K in this study, I’m also interested in how other lipid metabolizing enzymes are regulated in response to changes in membrane environments. If they are regulated similarly to PIP5K, those enzymes should have a lipid-sensing motif and detect a unique membrane environment that is suitable for their enzymatic reaction. I would like to validate this possibility in the near future. In addition, I’m curious about how heterogeneous lipid environments are maintained in cells and how this is conserved across species. Although information about lipid distribution is not ‘obviously’ coded in the genome, unique lipid environments of individual organelle compartments are highly conserved from yeast to mammals. This cannot be spontaneously created or explained by just biophysical properties of lipids alone. Therefore, I’m now thinking that some key information about lipid environments might be quietly coded in the genome.
Written by Taki Nishimura
 Close
Close