Accelerating antibody-based medicine development
Research at UCL has led to creation of web-based computational tools to help pharmaceutical companies develop antibody-based drugs more efficiently, resulting in over 200 patent applications.
12 April 2022
Antibodies form a critical part of the body’s immune response to infection. Scientists are exploiting their unique properties to develop new medicines; indeed, antibody-based drugs make up a third of all pharmaceuticals in development.
Antibodies have a unique Y-shape that allows them to ‘lock-on’ to ‘antigens’ – substances (mostly proteins) that the immune system has recognised as foreign. A third of all new medicines under development are based on antibodies, with around 100 licensed treatments for cancer, autoimmune disease and blood clotting, amongst others, and over 200 in development for COVID-19 alone.
Analysis of antibody sequences and structures
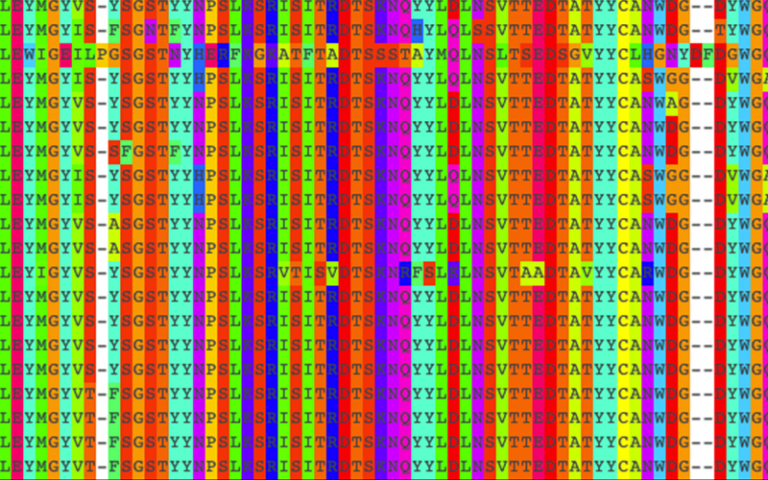
In 2008, Professor Andrew Martin’s team at the UCL Division of Biosciences developed the first automated method for applying standard numbering schemes to antibody sequences. Standard numbering allows different antibody sequences to be compared with one another and look for trends in the amino acids present at each position.
This method was developed into a tool called abYsis that could be used to analyse the amino acid sequences and 3D structures of antibodies and aid in modifying them for use as therapeutics. The UCL team went on to develop software to predict the antibodies’ structures, allowing key features to be highlighted in abYsis in the context of protein structure.
Reducing the risk of rejection
Antibody-based medicines can be rejected by the patient’s immune system if it recognises these antibody proteins as ‘foreign’. To mitigate this, abYsis allows investigators to assess how similar a particular antibody would be to human antibodies and to identify unusual features in their sequences. This has enabled pharmaceutical companies to reduce the risk of new antibody-based drugs not being tolerated and thus accelerating the drug design process.
Hundreds of biotechnology and pharmaceutical companies have used the software online in their drug design work and, since 2013, more than 200 patents applications have cited abYsis. Since its release in 2009, abYsis has undergone substantial development, including a new interface released in 2017.
A clinical and commercial success
Although abYsis is freely accessible online, commercial users can purchase a licence that allows them to store and analyse proprietary sequence and structure data locally without the risk of sending their data across the internet. Since 2013, seven major pharmaceutical companies have bought commercial licences for the software together with more than 20 smaller biotechnology companies. Commercial licences bought by these companies generated £325,000 in revenue in 2019/20.
In addition, patent lawyers, courts and the WHO are using the evidence and analysis provided by the software and presented by Professor Martin, to resolve patent disputes and standardise naming and annotation of antibody-based drugs.
Research synopsis
Antibody sequence and structure analysis to assist biologic drug design
Novel antibodies represent approximately a third of all drugs in development. Research at UCL has led to creation of free, web-based computational tools to help pharmaceutical companies develop antibody-based drugs. Since 2013, ‘abYsis’ which allows investigators to analyze the features of novel antibodies has been accessed by more than 44,000 unique users, resulting in over 200 patent applications. Commercial licences, bought by large pharmaceutical companies and small biotechnology companies, generated revenues over GBP325,000 in 2019/20. The software is being used to settle patent disputes at the European Patent Commission and in one case, Professor Martin’s expert testimony saved Genentech over GBP600,000. His expertise also informs naming and annotation of antibody-based drugs for the World Health Organisation.
Links
- Professor Andrew Martin’s academic profile
- UCL Division of Biosciences
- UCL Faculty of Life Sciences
- UCL Faculty of Life Sciences REF 2021
Image
- Image credit: abYsis
 Close
Close

