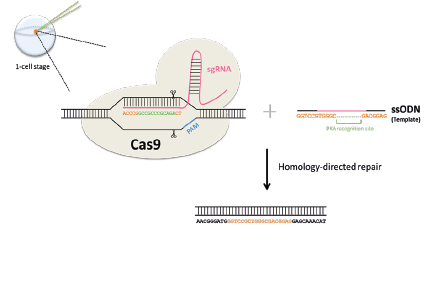
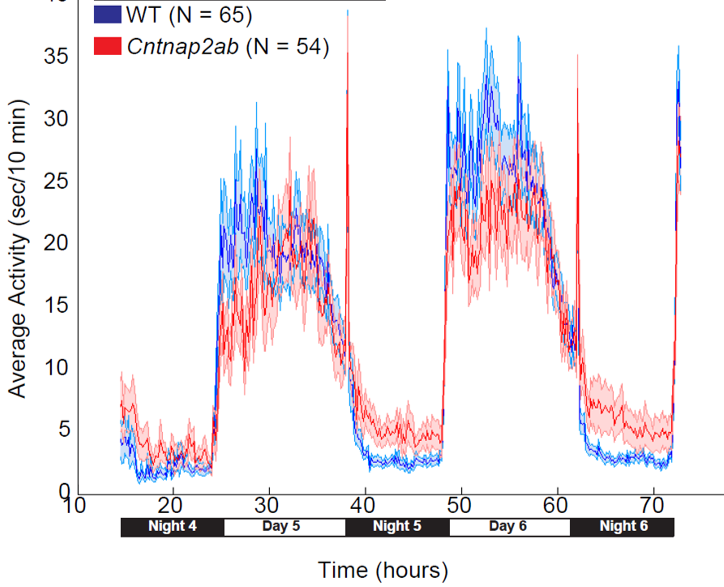
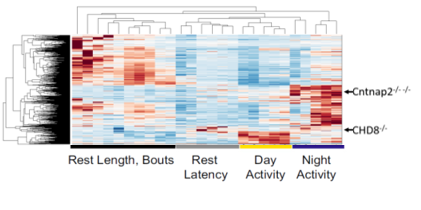
Making Sleep Mutants. In zebrafish, it is possible to quickly target specific genes to make mutants using a system called CRISPR-Cas9. This is done by injecting into 1-cell stage zebrafish eggs a Cas9 protein along with a single guide RNA (sgRNA) that has a complimentary sequence to the gene to target. The sgRNA will direct the Cas9 to cut the DNA at the gene of interest to create a double-strand break. When the cell repairs the break, it will occasionally make an error, leading to a mutation. If we supply a single stranded DNA oligonucleotide (ssODN) with homology to our targeted sequence, we can trick the cell’s repair machinery to replace the sequence with a specially edited sequence, for example one that is missing a few bases (green).